
Mycetocola lacteus
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales;
Average proteome isoelectric point is 6.18
Get precalculated fractions of proteins
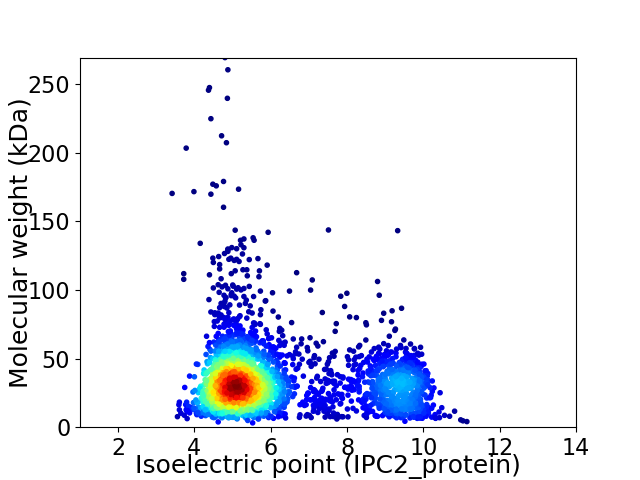
Virtual 2D-PAGE plot for 3070 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3L7ALP2|A0A3L7ALP2_9MICO Uncharacterized protein OS=Mycetocola lacteus OX=76637 GN=D9V34_11970 PE=4 SV=1
MM1 pKa = 7.24TGEE4 pKa = 4.2ALEE7 pKa = 4.58LANSTRR13 pKa = 11.84LGTVARR19 pKa = 11.84RR20 pKa = 11.84GLLAATLAVGLIIPALVPTAAFAAASATPGSVGQADD56 pKa = 3.88GTVTVSIAARR66 pKa = 11.84AAQLTLTPPSGVLIQRR82 pKa = 11.84PPTSFPCYY90 pKa = 9.47DD91 pKa = 3.39TGSGNTLIPASSTAFYY107 pKa = 9.97TGIYY111 pKa = 9.29CGYY114 pKa = 10.11SDD116 pKa = 3.8NTVGDD121 pKa = 3.78FTSYY125 pKa = 10.85AFAYY129 pKa = 9.18KK130 pKa = 10.22VSEE133 pKa = 4.02NAVPGSALGNFEE145 pKa = 4.09YY146 pKa = 10.42SVRR149 pKa = 11.84IPGSPNQTASAPLSVKK165 pKa = 9.56TLTAPTIATPTAGQLTNNPRR185 pKa = 11.84PVISGTADD193 pKa = 3.39NLPGTTLAVTITDD206 pKa = 3.59ADD208 pKa = 3.78TGVALAEE215 pKa = 4.16VPVVNGAWTYY225 pKa = 10.08TPVANRR231 pKa = 11.84PEE233 pKa = 4.21GTNNISVAASLNGVTTTATTRR254 pKa = 11.84SFVVDD259 pKa = 4.82TIPPAVPVLTSPVAGSATANTTPTVSGTGEE289 pKa = 3.72NGSRR293 pKa = 11.84IEE295 pKa = 3.91VRR297 pKa = 11.84NDD299 pKa = 2.76AGAVLCEE306 pKa = 4.01ATVVAGAWSCVVAPALTEE324 pKa = 4.38GSHH327 pKa = 5.78TLTPVALDD335 pKa = 3.66EE336 pKa = 5.24AGNAAPGTPVTFSVITTPPTAPVITAPTTGTITNNKK372 pKa = 7.95TPLITGTADD381 pKa = 2.58SGTIVEE387 pKa = 4.66IQNAANQLLCSGDD400 pKa = 3.55VPEE403 pKa = 5.99GEE405 pKa = 4.65FEE407 pKa = 4.58CVVAPALVDD416 pKa = 4.02GSHH419 pKa = 6.53TLTPISVDD427 pKa = 2.86AAGNRR432 pKa = 11.84TPGTPITIVVDD443 pKa = 4.21TVPPAAPVISSPATGTITKK462 pKa = 7.74NTRR465 pKa = 11.84PTFTGTGEE473 pKa = 3.88TGSRR477 pKa = 11.84VEE479 pKa = 3.49IHH481 pKa = 6.49GAQGQILCTAVVTAGAWSCQPATALEE507 pKa = 4.63DD508 pKa = 3.94GEE510 pKa = 4.67HH511 pKa = 6.97ALTAIAVDD519 pKa = 3.53TAGNRR524 pKa = 11.84TTGTSITITIDD535 pKa = 2.99TVAPTTPVITAPADD549 pKa = 3.49GTLTNNATPTITGTGEE565 pKa = 3.74NGSVIEE571 pKa = 4.38VRR573 pKa = 11.84GAEE576 pKa = 3.95NALLCTAVVAAGTWSCEE593 pKa = 3.53PATALGEE600 pKa = 4.41GSHH603 pKa = 6.25TLTPTARR610 pKa = 11.84DD611 pKa = 3.38AAGNTAPGTAVTITIDD627 pKa = 3.67TTPPAAPVITAPGDD641 pKa = 3.59GALLTTTTPTITGTGEE657 pKa = 3.69NGSTVEE663 pKa = 3.86VRR665 pKa = 11.84GGNNEE670 pKa = 4.06VLCTAVVTAGAWSCQISPALGQGDD694 pKa = 4.01HH695 pKa = 6.72SLTPVAMDD703 pKa = 3.73PAGNSTPGTAITVTVDD719 pKa = 3.29TVPPEE724 pKa = 4.22VPVITAPQNGTITNNATPTISGTGEE749 pKa = 3.69NGSTVEE755 pKa = 4.05VRR757 pKa = 11.84DD758 pKa = 3.51GAGTVLCTATVAAGIWSCTLDD779 pKa = 3.52EE780 pKa = 5.05PLTEE784 pKa = 4.97GDD786 pKa = 4.09HH787 pKa = 6.68PLLPVARR794 pKa = 11.84DD795 pKa = 3.32AAGNEE800 pKa = 4.08APGTAVTVTIDD811 pKa = 3.94LTPPAAPVVTAPLTGTATNVSTPVFTGTGEE841 pKa = 3.96NGSTVEE847 pKa = 4.11IRR849 pKa = 11.84AADD852 pKa = 3.77GTVLCSSLVEE862 pKa = 4.54DD863 pKa = 5.12GAWTCTIAPALDD875 pKa = 3.55EE876 pKa = 4.49GMHH879 pKa = 6.4ILSAVAVDD887 pKa = 3.43AAGNQTPGTIVALTVDD903 pKa = 4.28ITPPAVPAITSPVSGTITNDD923 pKa = 3.25TTPTLSGSGEE933 pKa = 4.08TGSTVEE939 pKa = 3.69IHH941 pKa = 5.85NAAGEE946 pKa = 4.4VLCSAVVTAGAWTCDD961 pKa = 3.51LTPALAEE968 pKa = 4.37GDD970 pKa = 3.91HH971 pKa = 6.29VLTPVAKK978 pKa = 10.31DD979 pKa = 3.15AAGNRR984 pKa = 11.84ATGLPITITVDD995 pKa = 3.52LTPPAVPVITSPVDD1009 pKa = 3.36GFVTNDD1015 pKa = 3.35TTPTLSGTGEE1025 pKa = 4.0TGSTVEE1031 pKa = 4.15IQNAAGDD1038 pKa = 4.21VLCSTVVTAGAWSCDD1053 pKa = 2.4ITTALTEE1060 pKa = 4.45GDD1062 pKa = 3.77HH1063 pKa = 6.67VLTPVAKK1070 pKa = 10.29DD1071 pKa = 3.02AAGNKK1076 pKa = 8.75TSGTAITITVDD1087 pKa = 3.08VTAPAVPVITSPVDD1101 pKa = 3.36GFVTNDD1107 pKa = 3.35TTPTLSGTGEE1117 pKa = 4.0TGSTVEE1123 pKa = 4.93IRR1125 pKa = 11.84NADD1128 pKa = 3.83DD1129 pKa = 3.91QVLCSAVVTAGAWTCDD1145 pKa = 3.56LAPALGDD1152 pKa = 3.7GNHH1155 pKa = 5.93VLTPVAKK1162 pKa = 10.29DD1163 pKa = 3.0AAGNKK1168 pKa = 8.65TPGTAITIEE1177 pKa = 4.68VITEE1181 pKa = 3.97LPGAPIVTSPTNGTITNDD1199 pKa = 3.09ATPTLTGTGEE1209 pKa = 3.96NGTTVEE1215 pKa = 4.15VRR1217 pKa = 11.84DD1218 pKa = 3.53AAGVVLCSAPVTDD1231 pKa = 4.77GTWSCVVAPALTDD1244 pKa = 3.73GDD1246 pKa = 4.36HH1247 pKa = 6.65EE1248 pKa = 4.46LVAVAVDD1255 pKa = 3.44AASNEE1260 pKa = 4.43TPGIPLTITVDD1271 pKa = 3.73TVPPAVPVITAPLDD1285 pKa = 3.55GTVTNVEE1292 pKa = 4.38SPTLSGSGEE1301 pKa = 4.11SGSTVEE1307 pKa = 4.25IQNAAGEE1314 pKa = 4.51VLCSVVVTAGAWSCEE1329 pKa = 3.87LAPALTEE1336 pKa = 4.3GDD1338 pKa = 3.73HH1339 pKa = 6.71VLTPVAKK1346 pKa = 10.23DD1347 pKa = 3.04AAGNEE1352 pKa = 4.39TPGVAITITVDD1363 pKa = 3.16VTPPAVPAITSPADD1377 pKa = 3.33GFVTNDD1383 pKa = 3.33TTPTLSGTGEE1393 pKa = 4.0TGSTVEE1399 pKa = 3.69IHH1401 pKa = 5.85NAAGEE1406 pKa = 4.4VLCSAVVTAGAWTCDD1421 pKa = 3.17LATTLTEE1428 pKa = 4.34GDD1430 pKa = 3.76HH1431 pKa = 6.55VLTPVAKK1438 pKa = 10.29DD1439 pKa = 3.0AAGNKK1444 pKa = 8.59TPGTPITITVDD1455 pKa = 3.18VTPPTVPVITSPANGLVTNDD1475 pKa = 3.53TTPTLSGTGEE1485 pKa = 4.05NGSTVTVKK1493 pKa = 10.51NAADD1497 pKa = 3.76EE1498 pKa = 4.78VVCSAVVSNGAWSCTLDD1515 pKa = 3.38SALTEE1520 pKa = 4.44GDD1522 pKa = 3.66HH1523 pKa = 6.73VLTPVATDD1531 pKa = 3.27AAGNKK1536 pKa = 8.25ATGTAITITVDD1547 pKa = 3.2TTAPAAPVIATPSANIVTNNTTPTLAGTGEE1577 pKa = 4.17TGSTVEE1583 pKa = 4.06IRR1585 pKa = 11.84NAAGVVLCSAEE1596 pKa = 4.24VVDD1599 pKa = 5.15GAWSCVVAPALTDD1612 pKa = 3.4GEE1614 pKa = 4.63YY1615 pKa = 10.4PLTAVAVDD1623 pKa = 3.27AAGNEE1628 pKa = 4.51TEE1630 pKa = 4.54GSSVTITVKK1639 pKa = 8.25TTLPAAPVITSPTNGSVGNNPAPTLSGTGEE1669 pKa = 4.01NGTTVEE1675 pKa = 3.89VRR1677 pKa = 11.84GPGGEE1682 pKa = 4.31VLCSAVVTNGTWSCEE1697 pKa = 3.86PSPEE1701 pKa = 4.1LTEE1704 pKa = 4.69GDD1706 pKa = 3.54HH1707 pKa = 6.35TLTPVAVDD1715 pKa = 3.0IAGNEE1720 pKa = 4.36TPGNPITLTVDD1731 pKa = 3.68TTPPTPTTDD1740 pKa = 3.35VVCSVNQLGEE1750 pKa = 4.38VTCEE1754 pKa = 3.65GTAAEE1759 pKa = 4.85GDD1761 pKa = 4.18TIVVTDD1767 pKa = 3.95GDD1769 pKa = 3.92GKK1771 pKa = 10.09EE1772 pKa = 3.84ICTVEE1777 pKa = 4.55VPAGGHH1783 pKa = 4.33WTCTGGPVTSFPVTVEE1799 pKa = 4.18TVDD1802 pKa = 3.39PAGNSGGVHH1811 pKa = 6.24TLPTPPIIVSPNTGSVLGEE1830 pKa = 4.07STPTLTGTGAAGDD1843 pKa = 4.03TVEE1846 pKa = 5.48LLDD1849 pKa = 4.15AEE1851 pKa = 4.64GSVVCTTTVEE1861 pKa = 4.68ADD1863 pKa = 3.92GTWSCTLPAALADD1876 pKa = 4.03GTHH1879 pKa = 6.52TLTPVATDD1887 pKa = 3.27ADD1889 pKa = 4.44GNTFTGIAISLNIDD1903 pKa = 3.46TVAPSPPTGVEE1914 pKa = 3.97CSANADD1920 pKa = 3.66GTVTCSGTAEE1930 pKa = 4.17PDD1932 pKa = 3.44STVEE1936 pKa = 3.76ITDD1939 pKa = 3.5PEE1941 pKa = 4.53GNLICEE1947 pKa = 4.5VTVGADD1953 pKa = 3.76GTWTCTSEE1961 pKa = 4.2GAVDD1965 pKa = 3.96AGEE1968 pKa = 4.04LTITVIDD1975 pKa = 3.81PAGNRR1980 pKa = 11.84SPSISIGVTPLPGTPGPGTTDD2001 pKa = 3.24PGAPGPGTPGPGTPGPGTPGPGTPGPGAGGGSLVTTGADD2040 pKa = 3.24AVGMGALALAAMLLLGSGVFLRR2062 pKa = 11.84SRR2064 pKa = 11.84RR2065 pKa = 11.84TSGTTRR2071 pKa = 11.84GEE2073 pKa = 4.19DD2074 pKa = 3.05
MM1 pKa = 7.24TGEE4 pKa = 4.2ALEE7 pKa = 4.58LANSTRR13 pKa = 11.84LGTVARR19 pKa = 11.84RR20 pKa = 11.84GLLAATLAVGLIIPALVPTAAFAAASATPGSVGQADD56 pKa = 3.88GTVTVSIAARR66 pKa = 11.84AAQLTLTPPSGVLIQRR82 pKa = 11.84PPTSFPCYY90 pKa = 9.47DD91 pKa = 3.39TGSGNTLIPASSTAFYY107 pKa = 9.97TGIYY111 pKa = 9.29CGYY114 pKa = 10.11SDD116 pKa = 3.8NTVGDD121 pKa = 3.78FTSYY125 pKa = 10.85AFAYY129 pKa = 9.18KK130 pKa = 10.22VSEE133 pKa = 4.02NAVPGSALGNFEE145 pKa = 4.09YY146 pKa = 10.42SVRR149 pKa = 11.84IPGSPNQTASAPLSVKK165 pKa = 9.56TLTAPTIATPTAGQLTNNPRR185 pKa = 11.84PVISGTADD193 pKa = 3.39NLPGTTLAVTITDD206 pKa = 3.59ADD208 pKa = 3.78TGVALAEE215 pKa = 4.16VPVVNGAWTYY225 pKa = 10.08TPVANRR231 pKa = 11.84PEE233 pKa = 4.21GTNNISVAASLNGVTTTATTRR254 pKa = 11.84SFVVDD259 pKa = 4.82TIPPAVPVLTSPVAGSATANTTPTVSGTGEE289 pKa = 3.72NGSRR293 pKa = 11.84IEE295 pKa = 3.91VRR297 pKa = 11.84NDD299 pKa = 2.76AGAVLCEE306 pKa = 4.01ATVVAGAWSCVVAPALTEE324 pKa = 4.38GSHH327 pKa = 5.78TLTPVALDD335 pKa = 3.66EE336 pKa = 5.24AGNAAPGTPVTFSVITTPPTAPVITAPTTGTITNNKK372 pKa = 7.95TPLITGTADD381 pKa = 2.58SGTIVEE387 pKa = 4.66IQNAANQLLCSGDD400 pKa = 3.55VPEE403 pKa = 5.99GEE405 pKa = 4.65FEE407 pKa = 4.58CVVAPALVDD416 pKa = 4.02GSHH419 pKa = 6.53TLTPISVDD427 pKa = 2.86AAGNRR432 pKa = 11.84TPGTPITIVVDD443 pKa = 4.21TVPPAAPVISSPATGTITKK462 pKa = 7.74NTRR465 pKa = 11.84PTFTGTGEE473 pKa = 3.88TGSRR477 pKa = 11.84VEE479 pKa = 3.49IHH481 pKa = 6.49GAQGQILCTAVVTAGAWSCQPATALEE507 pKa = 4.63DD508 pKa = 3.94GEE510 pKa = 4.67HH511 pKa = 6.97ALTAIAVDD519 pKa = 3.53TAGNRR524 pKa = 11.84TTGTSITITIDD535 pKa = 2.99TVAPTTPVITAPADD549 pKa = 3.49GTLTNNATPTITGTGEE565 pKa = 3.74NGSVIEE571 pKa = 4.38VRR573 pKa = 11.84GAEE576 pKa = 3.95NALLCTAVVAAGTWSCEE593 pKa = 3.53PATALGEE600 pKa = 4.41GSHH603 pKa = 6.25TLTPTARR610 pKa = 11.84DD611 pKa = 3.38AAGNTAPGTAVTITIDD627 pKa = 3.67TTPPAAPVITAPGDD641 pKa = 3.59GALLTTTTPTITGTGEE657 pKa = 3.69NGSTVEE663 pKa = 3.86VRR665 pKa = 11.84GGNNEE670 pKa = 4.06VLCTAVVTAGAWSCQISPALGQGDD694 pKa = 4.01HH695 pKa = 6.72SLTPVAMDD703 pKa = 3.73PAGNSTPGTAITVTVDD719 pKa = 3.29TVPPEE724 pKa = 4.22VPVITAPQNGTITNNATPTISGTGEE749 pKa = 3.69NGSTVEE755 pKa = 4.05VRR757 pKa = 11.84DD758 pKa = 3.51GAGTVLCTATVAAGIWSCTLDD779 pKa = 3.52EE780 pKa = 5.05PLTEE784 pKa = 4.97GDD786 pKa = 4.09HH787 pKa = 6.68PLLPVARR794 pKa = 11.84DD795 pKa = 3.32AAGNEE800 pKa = 4.08APGTAVTVTIDD811 pKa = 3.94LTPPAAPVVTAPLTGTATNVSTPVFTGTGEE841 pKa = 3.96NGSTVEE847 pKa = 4.11IRR849 pKa = 11.84AADD852 pKa = 3.77GTVLCSSLVEE862 pKa = 4.54DD863 pKa = 5.12GAWTCTIAPALDD875 pKa = 3.55EE876 pKa = 4.49GMHH879 pKa = 6.4ILSAVAVDD887 pKa = 3.43AAGNQTPGTIVALTVDD903 pKa = 4.28ITPPAVPAITSPVSGTITNDD923 pKa = 3.25TTPTLSGSGEE933 pKa = 4.08TGSTVEE939 pKa = 3.69IHH941 pKa = 5.85NAAGEE946 pKa = 4.4VLCSAVVTAGAWTCDD961 pKa = 3.51LTPALAEE968 pKa = 4.37GDD970 pKa = 3.91HH971 pKa = 6.29VLTPVAKK978 pKa = 10.31DD979 pKa = 3.15AAGNRR984 pKa = 11.84ATGLPITITVDD995 pKa = 3.52LTPPAVPVITSPVDD1009 pKa = 3.36GFVTNDD1015 pKa = 3.35TTPTLSGTGEE1025 pKa = 4.0TGSTVEE1031 pKa = 4.15IQNAAGDD1038 pKa = 4.21VLCSTVVTAGAWSCDD1053 pKa = 2.4ITTALTEE1060 pKa = 4.45GDD1062 pKa = 3.77HH1063 pKa = 6.67VLTPVAKK1070 pKa = 10.29DD1071 pKa = 3.02AAGNKK1076 pKa = 8.75TSGTAITITVDD1087 pKa = 3.08VTAPAVPVITSPVDD1101 pKa = 3.36GFVTNDD1107 pKa = 3.35TTPTLSGTGEE1117 pKa = 4.0TGSTVEE1123 pKa = 4.93IRR1125 pKa = 11.84NADD1128 pKa = 3.83DD1129 pKa = 3.91QVLCSAVVTAGAWTCDD1145 pKa = 3.56LAPALGDD1152 pKa = 3.7GNHH1155 pKa = 5.93VLTPVAKK1162 pKa = 10.29DD1163 pKa = 3.0AAGNKK1168 pKa = 8.65TPGTAITIEE1177 pKa = 4.68VITEE1181 pKa = 3.97LPGAPIVTSPTNGTITNDD1199 pKa = 3.09ATPTLTGTGEE1209 pKa = 3.96NGTTVEE1215 pKa = 4.15VRR1217 pKa = 11.84DD1218 pKa = 3.53AAGVVLCSAPVTDD1231 pKa = 4.77GTWSCVVAPALTDD1244 pKa = 3.73GDD1246 pKa = 4.36HH1247 pKa = 6.65EE1248 pKa = 4.46LVAVAVDD1255 pKa = 3.44AASNEE1260 pKa = 4.43TPGIPLTITVDD1271 pKa = 3.73TVPPAVPVITAPLDD1285 pKa = 3.55GTVTNVEE1292 pKa = 4.38SPTLSGSGEE1301 pKa = 4.11SGSTVEE1307 pKa = 4.25IQNAAGEE1314 pKa = 4.51VLCSVVVTAGAWSCEE1329 pKa = 3.87LAPALTEE1336 pKa = 4.3GDD1338 pKa = 3.73HH1339 pKa = 6.71VLTPVAKK1346 pKa = 10.23DD1347 pKa = 3.04AAGNEE1352 pKa = 4.39TPGVAITITVDD1363 pKa = 3.16VTPPAVPAITSPADD1377 pKa = 3.33GFVTNDD1383 pKa = 3.33TTPTLSGTGEE1393 pKa = 4.0TGSTVEE1399 pKa = 3.69IHH1401 pKa = 5.85NAAGEE1406 pKa = 4.4VLCSAVVTAGAWTCDD1421 pKa = 3.17LATTLTEE1428 pKa = 4.34GDD1430 pKa = 3.76HH1431 pKa = 6.55VLTPVAKK1438 pKa = 10.29DD1439 pKa = 3.0AAGNKK1444 pKa = 8.59TPGTPITITVDD1455 pKa = 3.18VTPPTVPVITSPANGLVTNDD1475 pKa = 3.53TTPTLSGTGEE1485 pKa = 4.05NGSTVTVKK1493 pKa = 10.51NAADD1497 pKa = 3.76EE1498 pKa = 4.78VVCSAVVSNGAWSCTLDD1515 pKa = 3.38SALTEE1520 pKa = 4.44GDD1522 pKa = 3.66HH1523 pKa = 6.73VLTPVATDD1531 pKa = 3.27AAGNKK1536 pKa = 8.25ATGTAITITVDD1547 pKa = 3.2TTAPAAPVIATPSANIVTNNTTPTLAGTGEE1577 pKa = 4.17TGSTVEE1583 pKa = 4.06IRR1585 pKa = 11.84NAAGVVLCSAEE1596 pKa = 4.24VVDD1599 pKa = 5.15GAWSCVVAPALTDD1612 pKa = 3.4GEE1614 pKa = 4.63YY1615 pKa = 10.4PLTAVAVDD1623 pKa = 3.27AAGNEE1628 pKa = 4.51TEE1630 pKa = 4.54GSSVTITVKK1639 pKa = 8.25TTLPAAPVITSPTNGSVGNNPAPTLSGTGEE1669 pKa = 4.01NGTTVEE1675 pKa = 3.89VRR1677 pKa = 11.84GPGGEE1682 pKa = 4.31VLCSAVVTNGTWSCEE1697 pKa = 3.86PSPEE1701 pKa = 4.1LTEE1704 pKa = 4.69GDD1706 pKa = 3.54HH1707 pKa = 6.35TLTPVAVDD1715 pKa = 3.0IAGNEE1720 pKa = 4.36TPGNPITLTVDD1731 pKa = 3.68TTPPTPTTDD1740 pKa = 3.35VVCSVNQLGEE1750 pKa = 4.38VTCEE1754 pKa = 3.65GTAAEE1759 pKa = 4.85GDD1761 pKa = 4.18TIVVTDD1767 pKa = 3.95GDD1769 pKa = 3.92GKK1771 pKa = 10.09EE1772 pKa = 3.84ICTVEE1777 pKa = 4.55VPAGGHH1783 pKa = 4.33WTCTGGPVTSFPVTVEE1799 pKa = 4.18TVDD1802 pKa = 3.39PAGNSGGVHH1811 pKa = 6.24TLPTPPIIVSPNTGSVLGEE1830 pKa = 4.07STPTLTGTGAAGDD1843 pKa = 4.03TVEE1846 pKa = 5.48LLDD1849 pKa = 4.15AEE1851 pKa = 4.64GSVVCTTTVEE1861 pKa = 4.68ADD1863 pKa = 3.92GTWSCTLPAALADD1876 pKa = 4.03GTHH1879 pKa = 6.52TLTPVATDD1887 pKa = 3.27ADD1889 pKa = 4.44GNTFTGIAISLNIDD1903 pKa = 3.46TVAPSPPTGVEE1914 pKa = 3.97CSANADD1920 pKa = 3.66GTVTCSGTAEE1930 pKa = 4.17PDD1932 pKa = 3.44STVEE1936 pKa = 3.76ITDD1939 pKa = 3.5PEE1941 pKa = 4.53GNLICEE1947 pKa = 4.5VTVGADD1953 pKa = 3.76GTWTCTSEE1961 pKa = 4.2GAVDD1965 pKa = 3.96AGEE1968 pKa = 4.04LTITVIDD1975 pKa = 3.81PAGNRR1980 pKa = 11.84SPSISIGVTPLPGTPGPGTTDD2001 pKa = 3.24PGAPGPGTPGPGTPGPGTPGPGTPGPGAGGGSLVTTGADD2040 pKa = 3.24AVGMGALALAAMLLLGSGVFLRR2062 pKa = 11.84SRR2064 pKa = 11.84RR2065 pKa = 11.84TSGTTRR2071 pKa = 11.84GEE2073 pKa = 4.19DD2074 pKa = 3.05
Molecular weight: 203.45 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3L7AHK3|A0A3L7AHK3_9MICO Succinate dehydrogenase flavoprotein subunit OS=Mycetocola lacteus OX=76637 GN=D9V34_15050 PE=3 SV=1
MM1 pKa = 7.4GSVIKK6 pKa = 10.38KK7 pKa = 8.42RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.06RR11 pKa = 11.84MSKK14 pKa = 9.76KK15 pKa = 9.54KK16 pKa = 9.79HH17 pKa = 5.67RR18 pKa = 11.84KK19 pKa = 8.31LLRR22 pKa = 11.84KK23 pKa = 7.78TRR25 pKa = 11.84HH26 pKa = 3.65QRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.78KK32 pKa = 9.85
MM1 pKa = 7.4GSVIKK6 pKa = 10.38KK7 pKa = 8.42RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.06RR11 pKa = 11.84MSKK14 pKa = 9.76KK15 pKa = 9.54KK16 pKa = 9.79HH17 pKa = 5.67RR18 pKa = 11.84KK19 pKa = 8.31LLRR22 pKa = 11.84KK23 pKa = 7.78TRR25 pKa = 11.84HH26 pKa = 3.65QRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.78KK32 pKa = 9.85
Molecular weight: 4.1 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1024074 |
29 |
2610 |
333.6 |
35.62 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.047 ± 0.071 | 0.465 ± 0.01 |
5.503 ± 0.035 | 5.609 ± 0.047 |
3.22 ± 0.028 | 8.972 ± 0.042 |
1.885 ± 0.02 | 5.228 ± 0.032 |
2.21 ± 0.042 | 10.351 ± 0.059 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.67 ± 0.018 | 2.493 ± 0.028 |
5.556 ± 0.039 | 2.775 ± 0.024 |
6.78 ± 0.05 | 5.92 ± 0.036 |
6.696 ± 0.054 | 8.263 ± 0.041 |
1.394 ± 0.018 | 1.964 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
