
Rosellinia necatrix partitivirus 2
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Duplopiviricetes; Durnavirales; Partitiviridae; Alphapartitivirus
Average proteome isoelectric point is 6.3
Get precalculated fractions of proteins
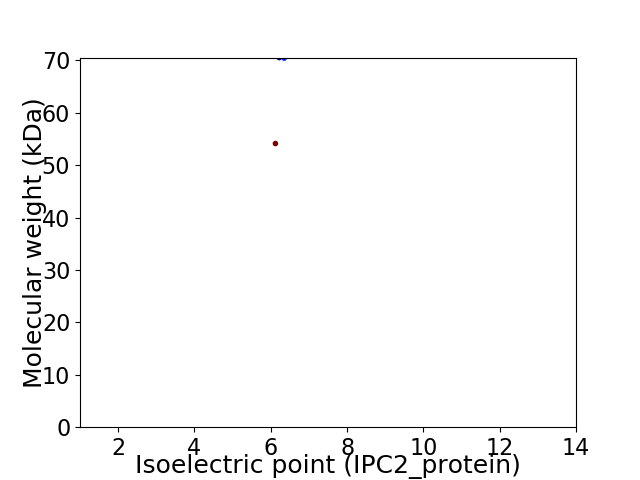
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|L8B8G3|L8B8G3_9VIRU RNA-dependent RNA polymerase OS=Rosellinia necatrix partitivirus 2 OX=859651 GN=RdRp PE=4 SV=1
MM1 pKa = 7.04STPSVTLSAFKK12 pKa = 10.67AAVAKK17 pKa = 10.67SGLEE21 pKa = 3.92DD22 pKa = 3.53QFLKK26 pKa = 10.2LTGLTSMDD34 pKa = 4.18EE35 pKa = 4.04LSADD39 pKa = 3.38NFNEE43 pKa = 4.08KK44 pKa = 10.2FGQADD49 pKa = 3.92SLTKK53 pKa = 10.63SILAPSAAPVPHH65 pKa = 6.22QEE67 pKa = 4.18QKK69 pKa = 10.96KK70 pKa = 9.56EE71 pKa = 3.78DD72 pKa = 3.78ADD74 pKa = 3.84AKK76 pKa = 10.39RR77 pKa = 11.84VSAPVSASDD86 pKa = 3.13FLYY89 pKa = 10.45GFKK92 pKa = 10.81HH93 pKa = 5.03MTIQYY98 pKa = 10.76ASRR101 pKa = 11.84KK102 pKa = 8.93KK103 pKa = 10.76NNTFLPSAFMMMYY116 pKa = 9.73IIHH119 pKa = 6.52EE120 pKa = 4.46MNVLLCEE127 pKa = 3.71HH128 pKa = 7.11FYY130 pKa = 10.84FKK132 pKa = 10.91RR133 pKa = 11.84EE134 pKa = 3.92APNYY138 pKa = 9.91HH139 pKa = 6.93PLLLRR144 pKa = 11.84TYY146 pKa = 10.29FGILFIIQTLRR157 pKa = 11.84AQHH160 pKa = 6.34AANNLFGDD168 pKa = 3.5QFEE171 pKa = 4.43FLEE174 pKa = 4.75KK175 pKa = 10.48FLDD178 pKa = 4.23AYY180 pKa = 9.99PVDD183 pKa = 4.2TLPIPAPLIHH193 pKa = 6.76IFKK196 pKa = 9.63SLCSSRR202 pKa = 11.84PEE204 pKa = 3.74IPQYY208 pKa = 11.3GYY210 pKa = 8.49VTPFVPEE217 pKa = 4.26EE218 pKa = 3.68LWGTQLKK225 pKa = 10.46DD226 pKa = 3.44SPANALYY233 pKa = 10.72ARR235 pKa = 11.84NFFPNIPYY243 pKa = 10.1ILSLYY248 pKa = 8.71ATLNSSTIKK257 pKa = 10.84GSVKK261 pKa = 10.29EE262 pKa = 4.08MNSYY266 pKa = 10.32LGNGTDD272 pKa = 3.82DD273 pKa = 3.39RR274 pKa = 11.84TFAGHH279 pKa = 7.5AYY281 pKa = 10.43DD282 pKa = 5.23KK283 pKa = 11.41DD284 pKa = 3.44PAQWTEE290 pKa = 3.44EE291 pKa = 3.79RR292 pKa = 11.84AFFLRR297 pKa = 11.84SPGMEE302 pKa = 3.86HH303 pKa = 6.94ALEE306 pKa = 4.61GTSSFNKK313 pKa = 10.3DD314 pKa = 2.89FFEE317 pKa = 4.4NRR319 pKa = 11.84FDD321 pKa = 3.81NLEE324 pKa = 3.99VPIPSTNDD332 pKa = 2.67KK333 pKa = 11.13VKK335 pKa = 10.58FVDD338 pKa = 3.53QFCYY342 pKa = 10.36LSKK345 pKa = 10.75NLAWFRR351 pKa = 11.84DD352 pKa = 3.65LVEE355 pKa = 4.4IASAAAKK362 pKa = 9.97FFEE365 pKa = 5.12GSGTLADD372 pKa = 4.68CSPVGPPVCQVQTTISNVDD391 pKa = 3.42LHH393 pKa = 6.07TAPTSISKK401 pKa = 10.6GKK403 pKa = 10.04SFYY406 pKa = 11.34AEE408 pKa = 3.42AHH410 pKa = 6.05MKK412 pKa = 9.01LTTTSRR418 pKa = 11.84TMDD421 pKa = 3.74QLSMIMASASHH432 pKa = 6.6LNLRR436 pKa = 11.84YY437 pKa = 9.91AHH439 pKa = 6.53ATHH442 pKa = 7.31SIQSRR447 pKa = 11.84TSGPFWSLNPIEE459 pKa = 5.3SSVTDD464 pKa = 3.98EE465 pKa = 4.22SSLIQVRR472 pKa = 11.84QTVKK476 pKa = 10.91KK477 pKa = 9.23MMKK480 pKa = 10.17SKK482 pKa = 10.74VV483 pKa = 3.28
MM1 pKa = 7.04STPSVTLSAFKK12 pKa = 10.67AAVAKK17 pKa = 10.67SGLEE21 pKa = 3.92DD22 pKa = 3.53QFLKK26 pKa = 10.2LTGLTSMDD34 pKa = 4.18EE35 pKa = 4.04LSADD39 pKa = 3.38NFNEE43 pKa = 4.08KK44 pKa = 10.2FGQADD49 pKa = 3.92SLTKK53 pKa = 10.63SILAPSAAPVPHH65 pKa = 6.22QEE67 pKa = 4.18QKK69 pKa = 10.96KK70 pKa = 9.56EE71 pKa = 3.78DD72 pKa = 3.78ADD74 pKa = 3.84AKK76 pKa = 10.39RR77 pKa = 11.84VSAPVSASDD86 pKa = 3.13FLYY89 pKa = 10.45GFKK92 pKa = 10.81HH93 pKa = 5.03MTIQYY98 pKa = 10.76ASRR101 pKa = 11.84KK102 pKa = 8.93KK103 pKa = 10.76NNTFLPSAFMMMYY116 pKa = 9.73IIHH119 pKa = 6.52EE120 pKa = 4.46MNVLLCEE127 pKa = 3.71HH128 pKa = 7.11FYY130 pKa = 10.84FKK132 pKa = 10.91RR133 pKa = 11.84EE134 pKa = 3.92APNYY138 pKa = 9.91HH139 pKa = 6.93PLLLRR144 pKa = 11.84TYY146 pKa = 10.29FGILFIIQTLRR157 pKa = 11.84AQHH160 pKa = 6.34AANNLFGDD168 pKa = 3.5QFEE171 pKa = 4.43FLEE174 pKa = 4.75KK175 pKa = 10.48FLDD178 pKa = 4.23AYY180 pKa = 9.99PVDD183 pKa = 4.2TLPIPAPLIHH193 pKa = 6.76IFKK196 pKa = 9.63SLCSSRR202 pKa = 11.84PEE204 pKa = 3.74IPQYY208 pKa = 11.3GYY210 pKa = 8.49VTPFVPEE217 pKa = 4.26EE218 pKa = 3.68LWGTQLKK225 pKa = 10.46DD226 pKa = 3.44SPANALYY233 pKa = 10.72ARR235 pKa = 11.84NFFPNIPYY243 pKa = 10.1ILSLYY248 pKa = 8.71ATLNSSTIKK257 pKa = 10.84GSVKK261 pKa = 10.29EE262 pKa = 4.08MNSYY266 pKa = 10.32LGNGTDD272 pKa = 3.82DD273 pKa = 3.39RR274 pKa = 11.84TFAGHH279 pKa = 7.5AYY281 pKa = 10.43DD282 pKa = 5.23KK283 pKa = 11.41DD284 pKa = 3.44PAQWTEE290 pKa = 3.44EE291 pKa = 3.79RR292 pKa = 11.84AFFLRR297 pKa = 11.84SPGMEE302 pKa = 3.86HH303 pKa = 6.94ALEE306 pKa = 4.61GTSSFNKK313 pKa = 10.3DD314 pKa = 2.89FFEE317 pKa = 4.4NRR319 pKa = 11.84FDD321 pKa = 3.81NLEE324 pKa = 3.99VPIPSTNDD332 pKa = 2.67KK333 pKa = 11.13VKK335 pKa = 10.58FVDD338 pKa = 3.53QFCYY342 pKa = 10.36LSKK345 pKa = 10.75NLAWFRR351 pKa = 11.84DD352 pKa = 3.65LVEE355 pKa = 4.4IASAAAKK362 pKa = 9.97FFEE365 pKa = 5.12GSGTLADD372 pKa = 4.68CSPVGPPVCQVQTTISNVDD391 pKa = 3.42LHH393 pKa = 6.07TAPTSISKK401 pKa = 10.6GKK403 pKa = 10.04SFYY406 pKa = 11.34AEE408 pKa = 3.42AHH410 pKa = 6.05MKK412 pKa = 9.01LTTTSRR418 pKa = 11.84TMDD421 pKa = 3.74QLSMIMASASHH432 pKa = 6.6LNLRR436 pKa = 11.84YY437 pKa = 9.91AHH439 pKa = 6.53ATHH442 pKa = 7.31SIQSRR447 pKa = 11.84TSGPFWSLNPIEE459 pKa = 5.3SSVTDD464 pKa = 3.98EE465 pKa = 4.22SSLIQVRR472 pKa = 11.84QTVKK476 pKa = 10.91KK477 pKa = 9.23MMKK480 pKa = 10.17SKK482 pKa = 10.74VV483 pKa = 3.28
Molecular weight: 54.12 kDa
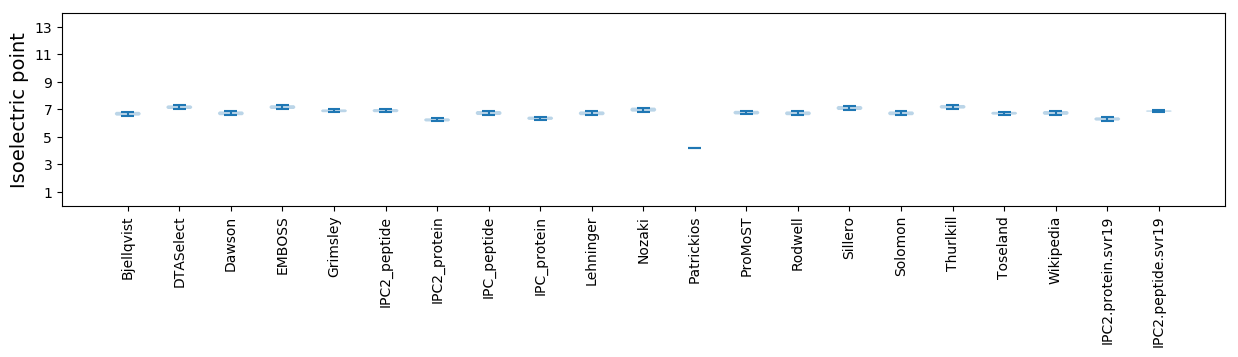
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|L8B8G3|L8B8G3_9VIRU RNA-dependent RNA polymerase OS=Rosellinia necatrix partitivirus 2 OX=859651 GN=RdRp PE=4 SV=1
MM1 pKa = 7.57KK2 pKa = 10.39NITDD6 pKa = 4.21LPFPSKK12 pKa = 10.54GVPQFRR18 pKa = 11.84DD19 pKa = 3.2ALPIHH24 pKa = 6.4GRR26 pKa = 11.84DD27 pKa = 3.4PEE29 pKa = 4.48VPNPVTDD36 pKa = 3.81ASNRR40 pKa = 11.84IIDD43 pKa = 3.72FALRR47 pKa = 11.84KK48 pKa = 9.67HH49 pKa = 5.35LTSDD53 pKa = 3.46EE54 pKa = 3.95FDD56 pKa = 3.29QVVNGYY62 pKa = 9.44RR63 pKa = 11.84RR64 pKa = 11.84SPWNEE69 pKa = 3.34DD70 pKa = 3.24ALNKK74 pKa = 10.49DD75 pKa = 3.32IEE77 pKa = 4.38KK78 pKa = 10.48LDD80 pKa = 3.9SDD82 pKa = 4.0EE83 pKa = 4.14HH84 pKa = 5.7TVIKK88 pKa = 10.1DD89 pKa = 3.11QHH91 pKa = 6.55YY92 pKa = 9.95EE93 pKa = 3.6NAIQHH98 pKa = 4.85VQKK101 pKa = 10.87LLTPEE106 pKa = 4.81KK107 pKa = 9.77PLQPVHH113 pKa = 6.42FADD116 pKa = 3.45LRR118 pKa = 11.84RR119 pKa = 11.84YY120 pKa = 9.02KK121 pKa = 9.91WRR123 pKa = 11.84LSTNIGAPFASSKK136 pKa = 10.0HH137 pKa = 3.74WQDD140 pKa = 3.24YY141 pKa = 9.83VKK143 pKa = 11.03AKK145 pKa = 10.38FNHH148 pKa = 5.76FRR150 pKa = 11.84DD151 pKa = 3.81GTPFEE156 pKa = 5.15NIAHH160 pKa = 6.99RR161 pKa = 11.84DD162 pKa = 3.69LFVEE166 pKa = 4.24AHH168 pKa = 7.09KK169 pKa = 10.74DD170 pKa = 3.7SQPLEE175 pKa = 3.92ITDD178 pKa = 4.3ARR180 pKa = 11.84MTKK183 pKa = 10.2HH184 pKa = 6.11NLYY187 pKa = 9.62TEE189 pKa = 4.45AFYY192 pKa = 11.22VSRR195 pKa = 11.84EE196 pKa = 4.03TIHH199 pKa = 6.95RR200 pKa = 11.84IKK202 pKa = 10.96DD203 pKa = 3.26GEE205 pKa = 4.31TTDD208 pKa = 5.02RR209 pKa = 11.84YY210 pKa = 11.34GNDD213 pKa = 2.54ARR215 pKa = 11.84YY216 pKa = 9.48WNTAFARR223 pKa = 11.84QHH225 pKa = 5.79LVKK228 pKa = 10.44ADD230 pKa = 3.74EE231 pKa = 4.48DD232 pKa = 4.15DD233 pKa = 3.54KK234 pKa = 11.81VRR236 pKa = 11.84LVFGAPFSLLCAEE249 pKa = 5.89LMFIWPLFIFLLSLKK264 pKa = 10.46GSTAFMLWTFEE275 pKa = 4.26TIIGGWYY282 pKa = 9.6RR283 pKa = 11.84LVNFFTTYY291 pKa = 11.19ALRR294 pKa = 11.84HH295 pKa = 4.97STVVTVDD302 pKa = 2.42WSGFDD307 pKa = 2.7RR308 pKa = 11.84YY309 pKa = 10.5ARR311 pKa = 11.84HH312 pKa = 5.35TVIKK316 pKa = 9.69DD317 pKa = 2.72IHH319 pKa = 6.87HH320 pKa = 6.72RR321 pKa = 11.84IIRR324 pKa = 11.84PMFDD328 pKa = 3.43FSHH331 pKa = 7.09GYY333 pKa = 9.97HH334 pKa = 6.63PTRR337 pKa = 11.84DD338 pKa = 3.71YY339 pKa = 11.54PDD341 pKa = 3.24TSKK344 pKa = 9.96TKK346 pKa = 10.49DD347 pKa = 3.38GQSNEE352 pKa = 3.59WRR354 pKa = 11.84ITNLWNWMTDD364 pKa = 3.61AILSTPLLLPNGRR377 pKa = 11.84FIRR380 pKa = 11.84FNHH383 pKa = 5.65SGIYY387 pKa = 9.67SGYY390 pKa = 9.65FQTQILDD397 pKa = 3.73SIYY400 pKa = 9.67NLVMIFTILSRR411 pKa = 11.84MGFDD415 pKa = 4.81LDD417 pKa = 3.44KK418 pKa = 11.28CVIKK422 pKa = 11.04VQGDD426 pKa = 3.43DD427 pKa = 4.1SIFMLLCCFIMISTSFFTLFKK448 pKa = 10.78YY449 pKa = 9.48YY450 pKa = 10.87AEE452 pKa = 4.53YY453 pKa = 11.21YY454 pKa = 10.57FGAKK458 pKa = 10.04LNEE461 pKa = 4.35KK462 pKa = 9.96KK463 pKa = 10.75SEE465 pKa = 3.97IRR467 pKa = 11.84PSLQDD472 pKa = 3.57AEE474 pKa = 4.34VLKK477 pKa = 10.69YY478 pKa = 10.43RR479 pKa = 11.84NRR481 pKa = 11.84NGIPYY486 pKa = 9.3RR487 pKa = 11.84DD488 pKa = 4.25RR489 pKa = 11.84ISLLAQLRR497 pKa = 11.84HH498 pKa = 5.62PEE500 pKa = 3.69RR501 pKa = 11.84RR502 pKa = 11.84TDD504 pKa = 3.35VDD506 pKa = 4.19SVAARR511 pKa = 11.84CIGIAYY517 pKa = 7.91AACGQDD523 pKa = 2.7ATVYY527 pKa = 9.32LICEE531 pKa = 4.7DD532 pKa = 3.28IYY534 pKa = 11.44NYY536 pKa = 10.27LVKK539 pKa = 10.49KK540 pKa = 10.07YY541 pKa = 9.71DD542 pKa = 2.93AHH544 pKa = 6.47ARR546 pKa = 11.84QGEE549 pKa = 4.26LDD551 pKa = 3.5FMFRR555 pKa = 11.84HH556 pKa = 6.29LEE558 pKa = 3.89LHH560 pKa = 6.25DD561 pKa = 4.23TVPSAATFPSWFDD574 pKa = 3.44TMAHH578 pKa = 5.88LTDD581 pKa = 4.47GPRR584 pKa = 11.84DD585 pKa = 3.74PVPSHH590 pKa = 6.49WPTDD594 pKa = 3.72YY595 pKa = 11.22FIGLPGRR602 pKa = 11.84LL603 pKa = 3.5
MM1 pKa = 7.57KK2 pKa = 10.39NITDD6 pKa = 4.21LPFPSKK12 pKa = 10.54GVPQFRR18 pKa = 11.84DD19 pKa = 3.2ALPIHH24 pKa = 6.4GRR26 pKa = 11.84DD27 pKa = 3.4PEE29 pKa = 4.48VPNPVTDD36 pKa = 3.81ASNRR40 pKa = 11.84IIDD43 pKa = 3.72FALRR47 pKa = 11.84KK48 pKa = 9.67HH49 pKa = 5.35LTSDD53 pKa = 3.46EE54 pKa = 3.95FDD56 pKa = 3.29QVVNGYY62 pKa = 9.44RR63 pKa = 11.84RR64 pKa = 11.84SPWNEE69 pKa = 3.34DD70 pKa = 3.24ALNKK74 pKa = 10.49DD75 pKa = 3.32IEE77 pKa = 4.38KK78 pKa = 10.48LDD80 pKa = 3.9SDD82 pKa = 4.0EE83 pKa = 4.14HH84 pKa = 5.7TVIKK88 pKa = 10.1DD89 pKa = 3.11QHH91 pKa = 6.55YY92 pKa = 9.95EE93 pKa = 3.6NAIQHH98 pKa = 4.85VQKK101 pKa = 10.87LLTPEE106 pKa = 4.81KK107 pKa = 9.77PLQPVHH113 pKa = 6.42FADD116 pKa = 3.45LRR118 pKa = 11.84RR119 pKa = 11.84YY120 pKa = 9.02KK121 pKa = 9.91WRR123 pKa = 11.84LSTNIGAPFASSKK136 pKa = 10.0HH137 pKa = 3.74WQDD140 pKa = 3.24YY141 pKa = 9.83VKK143 pKa = 11.03AKK145 pKa = 10.38FNHH148 pKa = 5.76FRR150 pKa = 11.84DD151 pKa = 3.81GTPFEE156 pKa = 5.15NIAHH160 pKa = 6.99RR161 pKa = 11.84DD162 pKa = 3.69LFVEE166 pKa = 4.24AHH168 pKa = 7.09KK169 pKa = 10.74DD170 pKa = 3.7SQPLEE175 pKa = 3.92ITDD178 pKa = 4.3ARR180 pKa = 11.84MTKK183 pKa = 10.2HH184 pKa = 6.11NLYY187 pKa = 9.62TEE189 pKa = 4.45AFYY192 pKa = 11.22VSRR195 pKa = 11.84EE196 pKa = 4.03TIHH199 pKa = 6.95RR200 pKa = 11.84IKK202 pKa = 10.96DD203 pKa = 3.26GEE205 pKa = 4.31TTDD208 pKa = 5.02RR209 pKa = 11.84YY210 pKa = 11.34GNDD213 pKa = 2.54ARR215 pKa = 11.84YY216 pKa = 9.48WNTAFARR223 pKa = 11.84QHH225 pKa = 5.79LVKK228 pKa = 10.44ADD230 pKa = 3.74EE231 pKa = 4.48DD232 pKa = 4.15DD233 pKa = 3.54KK234 pKa = 11.81VRR236 pKa = 11.84LVFGAPFSLLCAEE249 pKa = 5.89LMFIWPLFIFLLSLKK264 pKa = 10.46GSTAFMLWTFEE275 pKa = 4.26TIIGGWYY282 pKa = 9.6RR283 pKa = 11.84LVNFFTTYY291 pKa = 11.19ALRR294 pKa = 11.84HH295 pKa = 4.97STVVTVDD302 pKa = 2.42WSGFDD307 pKa = 2.7RR308 pKa = 11.84YY309 pKa = 10.5ARR311 pKa = 11.84HH312 pKa = 5.35TVIKK316 pKa = 9.69DD317 pKa = 2.72IHH319 pKa = 6.87HH320 pKa = 6.72RR321 pKa = 11.84IIRR324 pKa = 11.84PMFDD328 pKa = 3.43FSHH331 pKa = 7.09GYY333 pKa = 9.97HH334 pKa = 6.63PTRR337 pKa = 11.84DD338 pKa = 3.71YY339 pKa = 11.54PDD341 pKa = 3.24TSKK344 pKa = 9.96TKK346 pKa = 10.49DD347 pKa = 3.38GQSNEE352 pKa = 3.59WRR354 pKa = 11.84ITNLWNWMTDD364 pKa = 3.61AILSTPLLLPNGRR377 pKa = 11.84FIRR380 pKa = 11.84FNHH383 pKa = 5.65SGIYY387 pKa = 9.67SGYY390 pKa = 9.65FQTQILDD397 pKa = 3.73SIYY400 pKa = 9.67NLVMIFTILSRR411 pKa = 11.84MGFDD415 pKa = 4.81LDD417 pKa = 3.44KK418 pKa = 11.28CVIKK422 pKa = 11.04VQGDD426 pKa = 3.43DD427 pKa = 4.1SIFMLLCCFIMISTSFFTLFKK448 pKa = 10.78YY449 pKa = 9.48YY450 pKa = 10.87AEE452 pKa = 4.53YY453 pKa = 11.21YY454 pKa = 10.57FGAKK458 pKa = 10.04LNEE461 pKa = 4.35KK462 pKa = 9.96KK463 pKa = 10.75SEE465 pKa = 3.97IRR467 pKa = 11.84PSLQDD472 pKa = 3.57AEE474 pKa = 4.34VLKK477 pKa = 10.69YY478 pKa = 10.43RR479 pKa = 11.84NRR481 pKa = 11.84NGIPYY486 pKa = 9.3RR487 pKa = 11.84DD488 pKa = 4.25RR489 pKa = 11.84ISLLAQLRR497 pKa = 11.84HH498 pKa = 5.62PEE500 pKa = 3.69RR501 pKa = 11.84RR502 pKa = 11.84TDD504 pKa = 3.35VDD506 pKa = 4.19SVAARR511 pKa = 11.84CIGIAYY517 pKa = 7.91AACGQDD523 pKa = 2.7ATVYY527 pKa = 9.32LICEE531 pKa = 4.7DD532 pKa = 3.28IYY534 pKa = 11.44NYY536 pKa = 10.27LVKK539 pKa = 10.49KK540 pKa = 10.07YY541 pKa = 9.71DD542 pKa = 2.93AHH544 pKa = 6.47ARR546 pKa = 11.84QGEE549 pKa = 4.26LDD551 pKa = 3.5FMFRR555 pKa = 11.84HH556 pKa = 6.29LEE558 pKa = 3.89LHH560 pKa = 6.25DD561 pKa = 4.23TVPSAATFPSWFDD574 pKa = 3.44TMAHH578 pKa = 5.88LTDD581 pKa = 4.47GPRR584 pKa = 11.84DD585 pKa = 3.74PVPSHH590 pKa = 6.49WPTDD594 pKa = 3.72YY595 pKa = 11.22FIGLPGRR602 pKa = 11.84LL603 pKa = 3.5
Molecular weight: 70.42 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1086 |
483 |
603 |
543.0 |
62.27 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.459 ± 0.986 | 1.105 ± 0.048 |
6.814 ± 1.26 | 4.604 ± 0.391 |
6.814 ± 0.154 | 4.236 ± 0.206 |
3.683 ± 0.536 | 5.709 ± 0.788 |
5.525 ± 0.469 | 8.84 ± 0.184 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.486 ± 0.423 | 4.236 ± 0.218 |
5.525 ± 0.327 | 3.223 ± 0.344 |
5.341 ± 1.385 | 7.459 ± 1.693 |
6.538 ± 0.06 | 4.696 ± 0.096 |
1.565 ± 0.503 | 4.144 ± 0.426 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
