
Terracoccus luteus
Taxonomy: cellular organisms; Bacteria; Terrabacteria group;
Average proteome isoelectric point is 6.18
Get precalculated fractions of proteins
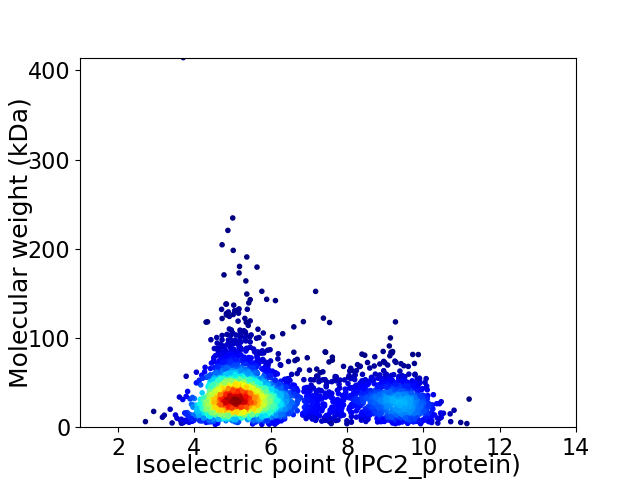
Virtual 2D-PAGE plot for 3555 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
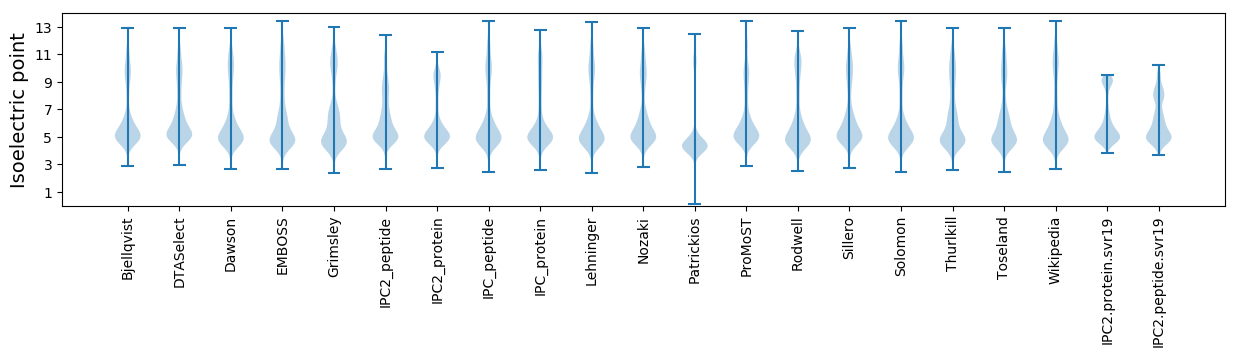
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A495Y1Q0|A0A495Y1Q0_9MICO L-lactate permease OS=Terracoccus luteus OX=53356 GN=DFJ68_2320 PE=3 SV=1
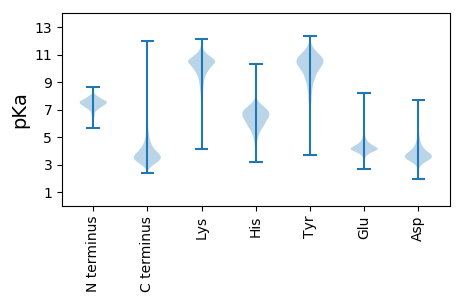
MM1 pKa = 7.44SLFITCPVEE10 pKa = 4.1SVEE13 pKa = 4.5RR14 pKa = 11.84ATAFYY19 pKa = 10.51TDD21 pKa = 5.27LGWTLNAEE29 pKa = 4.27MSDD32 pKa = 3.71HH33 pKa = 6.28NVSCFAIAPEE43 pKa = 4.12QYY45 pKa = 11.19VMLGSRR51 pKa = 11.84EE52 pKa = 4.01MYY54 pKa = 10.34ASVGGVEE61 pKa = 4.18QLVGDD66 pKa = 4.59ADD68 pKa = 4.17TPSKK72 pKa = 10.21VTVSFDD78 pKa = 3.2LGSRR82 pKa = 11.84EE83 pKa = 3.99AVDD86 pKa = 3.89ALVEE90 pKa = 4.07RR91 pKa = 11.84AGAAGGRR98 pKa = 11.84VGDD101 pKa = 3.62TDD103 pKa = 4.55EE104 pKa = 4.55YY105 pKa = 10.25PFMYY109 pKa = 9.89QRR111 pKa = 11.84QFDD114 pKa = 4.24DD115 pKa = 3.81LDD117 pKa = 4.38GYY119 pKa = 10.87HH120 pKa = 6.72YY121 pKa = 11.3SPFWMKK127 pKa = 10.69PDD129 pKa = 4.09GDD131 pKa = 3.87PTAA134 pKa = 4.95
MM1 pKa = 7.44SLFITCPVEE10 pKa = 4.1SVEE13 pKa = 4.5RR14 pKa = 11.84ATAFYY19 pKa = 10.51TDD21 pKa = 5.27LGWTLNAEE29 pKa = 4.27MSDD32 pKa = 3.71HH33 pKa = 6.28NVSCFAIAPEE43 pKa = 4.12QYY45 pKa = 11.19VMLGSRR51 pKa = 11.84EE52 pKa = 4.01MYY54 pKa = 10.34ASVGGVEE61 pKa = 4.18QLVGDD66 pKa = 4.59ADD68 pKa = 4.17TPSKK72 pKa = 10.21VTVSFDD78 pKa = 3.2LGSRR82 pKa = 11.84EE83 pKa = 3.99AVDD86 pKa = 3.89ALVEE90 pKa = 4.07RR91 pKa = 11.84AGAAGGRR98 pKa = 11.84VGDD101 pKa = 3.62TDD103 pKa = 4.55EE104 pKa = 4.55YY105 pKa = 10.25PFMYY109 pKa = 9.89QRR111 pKa = 11.84QFDD114 pKa = 4.24DD115 pKa = 3.81LDD117 pKa = 4.38GYY119 pKa = 10.87HH120 pKa = 6.72YY121 pKa = 11.3SPFWMKK127 pKa = 10.69PDD129 pKa = 4.09GDD131 pKa = 3.87PTAA134 pKa = 4.95
Molecular weight: 14.71 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A495XWW1|A0A495XWW1_9MICO Anti-sigma factor antagonist OS=Terracoccus luteus OX=53356 GN=DFJ68_2526 PE=3 SV=1
MM1 pKa = 7.18VPASPVPRR9 pKa = 11.84RR10 pKa = 11.84LLVRR14 pKa = 11.84RR15 pKa = 11.84DD16 pKa = 3.33RR17 pKa = 11.84VHH19 pKa = 6.62RR20 pKa = 11.84RR21 pKa = 11.84PVLPHH26 pKa = 4.66VRR28 pKa = 11.84LRR30 pKa = 11.84RR31 pKa = 11.84APRR34 pKa = 11.84VRR36 pKa = 11.84RR37 pKa = 11.84RR38 pKa = 11.84RR39 pKa = 11.84LPARR43 pKa = 11.84RR44 pKa = 11.84PPPEE48 pKa = 4.86RR49 pKa = 11.84PLPWRR54 pKa = 11.84ARR56 pKa = 11.84PGRR59 pKa = 11.84HH60 pKa = 5.75PRR62 pKa = 11.84PVRR65 pKa = 11.84PGRR68 pKa = 11.84RR69 pKa = 11.84PPRR72 pKa = 11.84RR73 pKa = 11.84ARR75 pKa = 11.84HH76 pKa = 5.31RR77 pKa = 11.84PARR80 pKa = 11.84PPPRR84 pKa = 11.84TPPRR88 pKa = 11.84HH89 pKa = 5.97PPPVVRR95 pKa = 11.84PLPHH99 pKa = 6.27ARR101 pKa = 11.84VRR103 pKa = 11.84PRR105 pKa = 11.84RR106 pKa = 11.84PLRR109 pKa = 11.84PARR112 pKa = 11.84RR113 pKa = 11.84RR114 pKa = 11.84PPRR117 pKa = 11.84VRR119 pKa = 11.84RR120 pKa = 11.84RR121 pKa = 11.84AARR124 pKa = 11.84PHH126 pKa = 6.41RR127 pKa = 11.84VPARR131 pKa = 11.84PSRR134 pKa = 11.84PRR136 pKa = 11.84RR137 pKa = 11.84RR138 pKa = 11.84LRR140 pKa = 11.84PRR142 pKa = 11.84CRR144 pKa = 11.84RR145 pKa = 11.84PPRR148 pKa = 11.84PRR150 pKa = 11.84PLPSGCRR157 pKa = 11.84APVTVASRR165 pKa = 11.84VPAYY169 pKa = 9.82RR170 pKa = 11.84RR171 pKa = 11.84RR172 pKa = 11.84PVRR175 pKa = 11.84PPVRR179 pKa = 11.84VSRR182 pKa = 11.84APATTRR188 pKa = 11.84SRR190 pKa = 11.84RR191 pKa = 11.84ARR193 pKa = 11.84EE194 pKa = 3.54WVRR197 pKa = 11.84PVRR200 pKa = 11.84PVPVATRR207 pKa = 11.84ARR209 pKa = 11.84DD210 pKa = 3.29RR211 pKa = 11.84ATTRR215 pKa = 11.84SARR218 pKa = 11.84TRR220 pKa = 11.84GCRR223 pKa = 11.84VRR225 pKa = 11.84SSGQGPPVPAAPVALVVPAGPVRR248 pKa = 11.84PAPAVPVRR256 pKa = 11.84VLPPVAPRR264 pKa = 11.84VPAVRR269 pKa = 11.84ARR271 pKa = 11.84ARR273 pKa = 11.84AA274 pKa = 3.28
MM1 pKa = 7.18VPASPVPRR9 pKa = 11.84RR10 pKa = 11.84LLVRR14 pKa = 11.84RR15 pKa = 11.84DD16 pKa = 3.33RR17 pKa = 11.84VHH19 pKa = 6.62RR20 pKa = 11.84RR21 pKa = 11.84PVLPHH26 pKa = 4.66VRR28 pKa = 11.84LRR30 pKa = 11.84RR31 pKa = 11.84APRR34 pKa = 11.84VRR36 pKa = 11.84RR37 pKa = 11.84RR38 pKa = 11.84RR39 pKa = 11.84LPARR43 pKa = 11.84RR44 pKa = 11.84PPPEE48 pKa = 4.86RR49 pKa = 11.84PLPWRR54 pKa = 11.84ARR56 pKa = 11.84PGRR59 pKa = 11.84HH60 pKa = 5.75PRR62 pKa = 11.84PVRR65 pKa = 11.84PGRR68 pKa = 11.84RR69 pKa = 11.84PPRR72 pKa = 11.84RR73 pKa = 11.84ARR75 pKa = 11.84HH76 pKa = 5.31RR77 pKa = 11.84PARR80 pKa = 11.84PPPRR84 pKa = 11.84TPPRR88 pKa = 11.84HH89 pKa = 5.97PPPVVRR95 pKa = 11.84PLPHH99 pKa = 6.27ARR101 pKa = 11.84VRR103 pKa = 11.84PRR105 pKa = 11.84RR106 pKa = 11.84PLRR109 pKa = 11.84PARR112 pKa = 11.84RR113 pKa = 11.84RR114 pKa = 11.84PPRR117 pKa = 11.84VRR119 pKa = 11.84RR120 pKa = 11.84RR121 pKa = 11.84AARR124 pKa = 11.84PHH126 pKa = 6.41RR127 pKa = 11.84VPARR131 pKa = 11.84PSRR134 pKa = 11.84PRR136 pKa = 11.84RR137 pKa = 11.84RR138 pKa = 11.84LRR140 pKa = 11.84PRR142 pKa = 11.84CRR144 pKa = 11.84RR145 pKa = 11.84PPRR148 pKa = 11.84PRR150 pKa = 11.84PLPSGCRR157 pKa = 11.84APVTVASRR165 pKa = 11.84VPAYY169 pKa = 9.82RR170 pKa = 11.84RR171 pKa = 11.84RR172 pKa = 11.84PVRR175 pKa = 11.84PPVRR179 pKa = 11.84VSRR182 pKa = 11.84APATTRR188 pKa = 11.84SRR190 pKa = 11.84RR191 pKa = 11.84ARR193 pKa = 11.84EE194 pKa = 3.54WVRR197 pKa = 11.84PVRR200 pKa = 11.84PVPVATRR207 pKa = 11.84ARR209 pKa = 11.84DD210 pKa = 3.29RR211 pKa = 11.84ATTRR215 pKa = 11.84SARR218 pKa = 11.84TRR220 pKa = 11.84GCRR223 pKa = 11.84VRR225 pKa = 11.84SSGQGPPVPAAPVALVVPAGPVRR248 pKa = 11.84PAPAVPVRR256 pKa = 11.84VLPPVAPRR264 pKa = 11.84VPAVRR269 pKa = 11.84ARR271 pKa = 11.84ARR273 pKa = 11.84AA274 pKa = 3.28
Molecular weight: 31.51 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1213893 |
30 |
4235 |
341.5 |
36.31 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.384 ± 0.047 | 0.642 ± 0.008 |
6.683 ± 0.036 | 5.229 ± 0.036 |
2.623 ± 0.022 | 9.609 ± 0.037 |
2.188 ± 0.023 | 2.838 ± 0.03 |
1.733 ± 0.028 | 9.957 ± 0.055 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.654 ± 0.016 | 1.598 ± 0.021 |
5.855 ± 0.031 | 2.644 ± 0.02 |
7.989 ± 0.044 | 5.477 ± 0.033 |
6.602 ± 0.058 | 9.988 ± 0.046 |
1.516 ± 0.021 | 1.794 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
