
Parasulfuritortus cantonensis
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Betaproteobacteria; Nitrosomonadales; Thiobacillaceae; Parasulfuritortus
Average proteome isoelectric point is 6.56
Get precalculated fractions of proteins
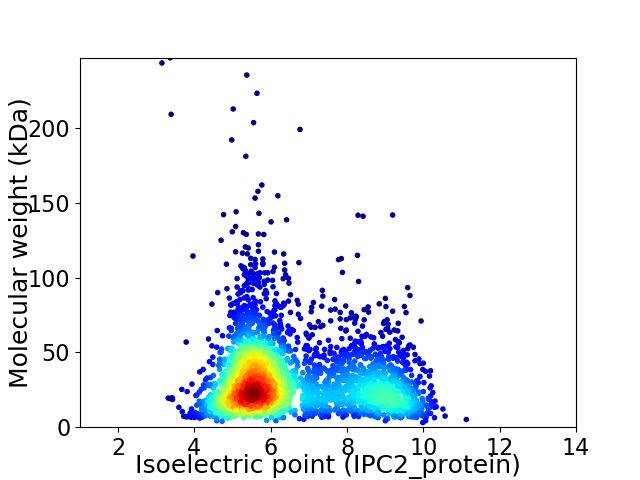
Virtual 2D-PAGE plot for 2975 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4V2NW73|A0A4V2NW73_9PROT Histidine kinase OS=Parasulfuritortus cantonensis OX=2528202 GN=EZJ19_05430 PE=4 SV=1
MM1 pKa = 7.2GTGLLNISLSGLNAAMAGIRR21 pKa = 11.84TVQQNIANANTAGYY35 pKa = 7.37TRR37 pKa = 11.84QEE39 pKa = 4.44VVQSASTPQYY49 pKa = 10.89SGGGYY54 pKa = 10.14LGTGVQVDD62 pKa = 4.18TVRR65 pKa = 11.84RR66 pKa = 11.84IYY68 pKa = 10.83DD69 pKa = 3.23QFLTSQTQNYY79 pKa = 6.08QTQLSAAEE87 pKa = 4.76AYY89 pKa = 10.12SSYY92 pKa = 11.05ASEE95 pKa = 3.93VDD97 pKa = 3.27ALLGGSSTGLSTPLQTFFSAVSEE120 pKa = 4.39VANDD124 pKa = 3.78PTSLTARR131 pKa = 11.84QQMISTANSLASRR144 pKa = 11.84FNLLADD150 pKa = 3.62NLQSIKK156 pKa = 10.17TEE158 pKa = 3.89INSEE162 pKa = 4.0VSTIATQINSYY173 pKa = 10.9ADD175 pKa = 3.43QIQSLNAEE183 pKa = 4.14IARR186 pKa = 11.84AQGSGQTPNDD196 pKa = 3.89LLDD199 pKa = 4.03QRR201 pKa = 11.84DD202 pKa = 3.66QLVNEE207 pKa = 4.61LGKK210 pKa = 10.54LVNVSSIQQSDD221 pKa = 3.6GTVAVMIGNGQPLVVGTAVRR241 pKa = 11.84HH242 pKa = 4.46VVAVSDD248 pKa = 4.31SADD251 pKa = 3.4PTQTILAMEE260 pKa = 4.93IPNSTATEE268 pKa = 4.25VIDD271 pKa = 4.25PSSITSGSLGGLFAFRR287 pKa = 11.84DD288 pKa = 3.84EE289 pKa = 4.34VLNPSIADD297 pKa = 4.02LDD299 pKa = 3.78QLAQSLADD307 pKa = 3.69AFNSQHH313 pKa = 5.77EE314 pKa = 4.26AGYY317 pKa = 10.46DD318 pKa = 3.48LNGAAGTAFFSYY330 pKa = 10.41SATSPAGTLAVVISQASQVAAASQGVSAAADD361 pKa = 3.52AGNSGDD367 pKa = 3.65ASIGAVTLSEE377 pKa = 4.37TVTRR381 pKa = 11.84PLADD385 pKa = 3.19APYY388 pKa = 8.54TLSYY392 pKa = 10.74NAGTVTVTDD401 pKa = 4.1ASANVVGSFTYY412 pKa = 10.32TSGATMTFDD421 pKa = 4.97GISVAITDD429 pKa = 4.07GSGGIADD436 pKa = 4.57GDD438 pKa = 4.0TFTIDD443 pKa = 3.05NSGAYY448 pKa = 9.57SGPGDD453 pKa = 3.77NGNALLLAGLQNEE466 pKa = 4.29AVIGSSTLSEE476 pKa = 4.46YY477 pKa = 9.63NTALVGRR484 pKa = 11.84NATYY488 pKa = 11.12ANSADD493 pKa = 3.75TNVSNFTSLYY503 pKa = 7.31QQSYY507 pKa = 11.39DD508 pKa = 3.44SLQSVSGVNLDD519 pKa = 3.63EE520 pKa = 4.59EE521 pKa = 4.55AVKK524 pKa = 10.7LIQLQQAYY532 pKa = 8.59QAAAKK537 pKa = 9.6AIQVSSTLFDD547 pKa = 3.61AVLGAMQQ554 pKa = 4.42
MM1 pKa = 7.2GTGLLNISLSGLNAAMAGIRR21 pKa = 11.84TVQQNIANANTAGYY35 pKa = 7.37TRR37 pKa = 11.84QEE39 pKa = 4.44VVQSASTPQYY49 pKa = 10.89SGGGYY54 pKa = 10.14LGTGVQVDD62 pKa = 4.18TVRR65 pKa = 11.84RR66 pKa = 11.84IYY68 pKa = 10.83DD69 pKa = 3.23QFLTSQTQNYY79 pKa = 6.08QTQLSAAEE87 pKa = 4.76AYY89 pKa = 10.12SSYY92 pKa = 11.05ASEE95 pKa = 3.93VDD97 pKa = 3.27ALLGGSSTGLSTPLQTFFSAVSEE120 pKa = 4.39VANDD124 pKa = 3.78PTSLTARR131 pKa = 11.84QQMISTANSLASRR144 pKa = 11.84FNLLADD150 pKa = 3.62NLQSIKK156 pKa = 10.17TEE158 pKa = 3.89INSEE162 pKa = 4.0VSTIATQINSYY173 pKa = 10.9ADD175 pKa = 3.43QIQSLNAEE183 pKa = 4.14IARR186 pKa = 11.84AQGSGQTPNDD196 pKa = 3.89LLDD199 pKa = 4.03QRR201 pKa = 11.84DD202 pKa = 3.66QLVNEE207 pKa = 4.61LGKK210 pKa = 10.54LVNVSSIQQSDD221 pKa = 3.6GTVAVMIGNGQPLVVGTAVRR241 pKa = 11.84HH242 pKa = 4.46VVAVSDD248 pKa = 4.31SADD251 pKa = 3.4PTQTILAMEE260 pKa = 4.93IPNSTATEE268 pKa = 4.25VIDD271 pKa = 4.25PSSITSGSLGGLFAFRR287 pKa = 11.84DD288 pKa = 3.84EE289 pKa = 4.34VLNPSIADD297 pKa = 4.02LDD299 pKa = 3.78QLAQSLADD307 pKa = 3.69AFNSQHH313 pKa = 5.77EE314 pKa = 4.26AGYY317 pKa = 10.46DD318 pKa = 3.48LNGAAGTAFFSYY330 pKa = 10.41SATSPAGTLAVVISQASQVAAASQGVSAAADD361 pKa = 3.52AGNSGDD367 pKa = 3.65ASIGAVTLSEE377 pKa = 4.37TVTRR381 pKa = 11.84PLADD385 pKa = 3.19APYY388 pKa = 8.54TLSYY392 pKa = 10.74NAGTVTVTDD401 pKa = 4.1ASANVVGSFTYY412 pKa = 10.32TSGATMTFDD421 pKa = 4.97GISVAITDD429 pKa = 4.07GSGGIADD436 pKa = 4.57GDD438 pKa = 4.0TFTIDD443 pKa = 3.05NSGAYY448 pKa = 9.57SGPGDD453 pKa = 3.77NGNALLLAGLQNEE466 pKa = 4.29AVIGSSTLSEE476 pKa = 4.46YY477 pKa = 9.63NTALVGRR484 pKa = 11.84NATYY488 pKa = 11.12ANSADD493 pKa = 3.75TNVSNFTSLYY503 pKa = 7.31QQSYY507 pKa = 11.39DD508 pKa = 3.44SLQSVSGVNLDD519 pKa = 3.63EE520 pKa = 4.59EE521 pKa = 4.55AVKK524 pKa = 10.7LIQLQQAYY532 pKa = 8.59QAAAKK537 pKa = 9.6AIQVSSTLFDD547 pKa = 3.61AVLGAMQQ554 pKa = 4.42
Molecular weight: 56.86 kDa
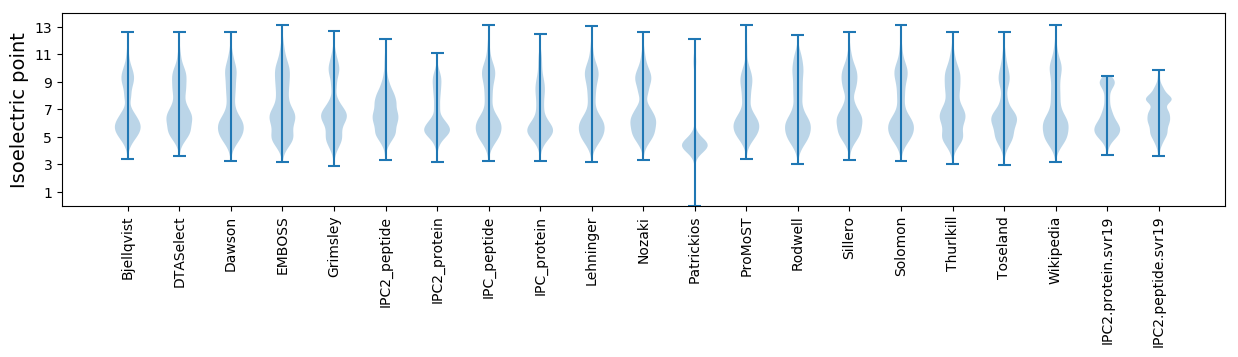
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4R1B8X1|A0A4R1B8X1_9PROT tRNA-2-methylthio-N(6)-dimethylallyladenosine synthase OS=Parasulfuritortus cantonensis OX=2528202 GN=miaB PE=3 SV=1
MM1 pKa = 7.35KK2 pKa = 9.36RR3 pKa = 11.84TYY5 pKa = 9.97QPSVVRR11 pKa = 11.84RR12 pKa = 11.84KK13 pKa = 8.57RR14 pKa = 11.84THH16 pKa = 5.82GFRR19 pKa = 11.84ARR21 pKa = 11.84MKK23 pKa = 8.8TKK25 pKa = 10.36GGRR28 pKa = 11.84AVINARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.58GRR39 pKa = 11.84KK40 pKa = 8.75RR41 pKa = 11.84LAVV44 pKa = 3.41
MM1 pKa = 7.35KK2 pKa = 9.36RR3 pKa = 11.84TYY5 pKa = 9.97QPSVVRR11 pKa = 11.84RR12 pKa = 11.84KK13 pKa = 8.57RR14 pKa = 11.84THH16 pKa = 5.82GFRR19 pKa = 11.84ARR21 pKa = 11.84MKK23 pKa = 8.8TKK25 pKa = 10.36GGRR28 pKa = 11.84AVINARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.58GRR39 pKa = 11.84KK40 pKa = 8.75RR41 pKa = 11.84LAVV44 pKa = 3.41
Molecular weight: 5.15 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
945077 |
26 |
2409 |
317.7 |
34.61 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.877 ± 0.079 | 0.979 ± 0.019 |
5.814 ± 0.042 | 5.712 ± 0.044 |
3.417 ± 0.027 | 8.463 ± 0.043 |
2.308 ± 0.023 | 4.268 ± 0.035 |
3.233 ± 0.042 | 11.139 ± 0.062 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.397 ± 0.021 | 2.603 ± 0.029 |
5.143 ± 0.034 | 3.333 ± 0.026 |
7.364 ± 0.051 | 4.719 ± 0.034 |
4.677 ± 0.052 | 7.538 ± 0.038 |
1.419 ± 0.02 | 2.595 ± 0.025 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
