
Methylobacterium frigidaeris
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Methylobacteriaceae; Methylobacterium
Average proteome isoelectric point is 6.89
Get precalculated fractions of proteins
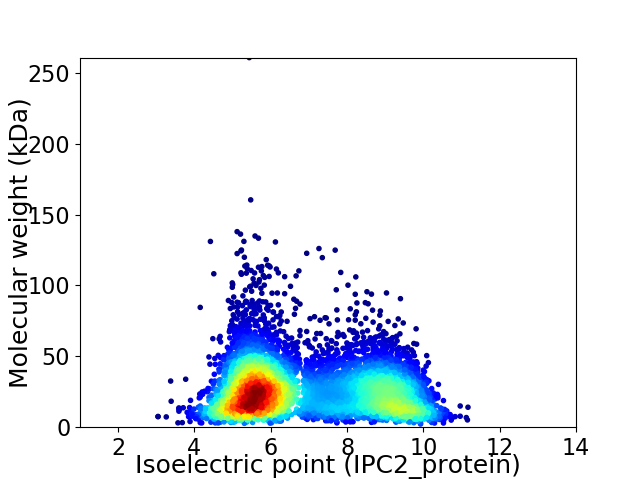
Virtual 2D-PAGE plot for 6432 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2G8MQU0|A0A2G8MQU0_9RHIZ Adenylosuccinate synthase OS=Methylobacterium frigidaeris OX=2038277 GN=CS379_00435 PE=4 SV=1
MM1 pKa = 7.47LSAALRR7 pKa = 11.84ALGCLTADD15 pKa = 3.55RR16 pKa = 11.84VLPTADD22 pKa = 3.98LGTVTVDD29 pKa = 3.78TVPDD33 pKa = 3.66GCGPGWPCEE42 pKa = 4.15ALPGTPPSVADD53 pKa = 4.02RR54 pKa = 11.84QAAPSADD61 pKa = 3.43DD62 pKa = 5.37LYY64 pKa = 10.52PQIAGVLTPRR74 pKa = 11.84GPAWGTDD81 pKa = 3.18EE82 pKa = 5.98AGDD85 pKa = 3.84GRR87 pKa = 11.84GAGPVMRR94 pKa = 11.84GVWRR98 pKa = 11.84ALSQWVARR106 pKa = 11.84QNADD110 pKa = 2.54EE111 pKa = 4.31WTLATQALPSAITYY125 pKa = 8.4SLPDD129 pKa = 3.22WEE131 pKa = 5.2AEE133 pKa = 4.04YY134 pKa = 11.06GLPDD138 pKa = 3.9PCSPGGFTTEE148 pKa = 3.51SRR150 pKa = 11.84IAAVRR155 pKa = 11.84ARR157 pKa = 11.84FGARR161 pKa = 11.84GGSSPAYY168 pKa = 8.78FICLAASLGYY178 pKa = 10.38AITIEE183 pKa = 4.43EE184 pKa = 4.42PTQFLCDD191 pKa = 3.47VSAVGTDD198 pKa = 3.3ALVEE202 pKa = 4.15TWFTCDD208 pKa = 4.53GGEE211 pKa = 4.33CDD213 pKa = 4.41GDD215 pKa = 4.34PIEE218 pKa = 4.86GFAMAPAADD227 pKa = 4.49DD228 pKa = 3.66TDD230 pKa = 3.94QVGGDD235 pKa = 3.65PFVEE239 pKa = 5.16GWCVADD245 pKa = 5.3EE246 pKa = 4.66GMCDD250 pKa = 3.4VTPLEE255 pKa = 5.04FYY257 pKa = 9.37ATNPNADD264 pKa = 2.95AWQFWVVHH272 pKa = 5.64VAAQGDD278 pKa = 3.76TWFACDD284 pKa = 4.76EE285 pKa = 4.71GEE287 pKa = 5.06CDD289 pKa = 3.48TDD291 pKa = 4.4PIEE294 pKa = 4.65GFRR297 pKa = 11.84SAADD301 pKa = 3.73LEE303 pKa = 4.47CLLRR307 pKa = 11.84RR308 pKa = 11.84VSPAHH313 pKa = 5.38TALVIAYY320 pKa = 9.24DD321 pKa = 3.75GG322 pKa = 3.79
MM1 pKa = 7.47LSAALRR7 pKa = 11.84ALGCLTADD15 pKa = 3.55RR16 pKa = 11.84VLPTADD22 pKa = 3.98LGTVTVDD29 pKa = 3.78TVPDD33 pKa = 3.66GCGPGWPCEE42 pKa = 4.15ALPGTPPSVADD53 pKa = 4.02RR54 pKa = 11.84QAAPSADD61 pKa = 3.43DD62 pKa = 5.37LYY64 pKa = 10.52PQIAGVLTPRR74 pKa = 11.84GPAWGTDD81 pKa = 3.18EE82 pKa = 5.98AGDD85 pKa = 3.84GRR87 pKa = 11.84GAGPVMRR94 pKa = 11.84GVWRR98 pKa = 11.84ALSQWVARR106 pKa = 11.84QNADD110 pKa = 2.54EE111 pKa = 4.31WTLATQALPSAITYY125 pKa = 8.4SLPDD129 pKa = 3.22WEE131 pKa = 5.2AEE133 pKa = 4.04YY134 pKa = 11.06GLPDD138 pKa = 3.9PCSPGGFTTEE148 pKa = 3.51SRR150 pKa = 11.84IAAVRR155 pKa = 11.84ARR157 pKa = 11.84FGARR161 pKa = 11.84GGSSPAYY168 pKa = 8.78FICLAASLGYY178 pKa = 10.38AITIEE183 pKa = 4.43EE184 pKa = 4.42PTQFLCDD191 pKa = 3.47VSAVGTDD198 pKa = 3.3ALVEE202 pKa = 4.15TWFTCDD208 pKa = 4.53GGEE211 pKa = 4.33CDD213 pKa = 4.41GDD215 pKa = 4.34PIEE218 pKa = 4.86GFAMAPAADD227 pKa = 4.49DD228 pKa = 3.66TDD230 pKa = 3.94QVGGDD235 pKa = 3.65PFVEE239 pKa = 5.16GWCVADD245 pKa = 5.3EE246 pKa = 4.66GMCDD250 pKa = 3.4VTPLEE255 pKa = 5.04FYY257 pKa = 9.37ATNPNADD264 pKa = 2.95AWQFWVVHH272 pKa = 5.64VAAQGDD278 pKa = 3.76TWFACDD284 pKa = 4.76EE285 pKa = 4.71GEE287 pKa = 5.06CDD289 pKa = 3.48TDD291 pKa = 4.4PIEE294 pKa = 4.65GFRR297 pKa = 11.84SAADD301 pKa = 3.73LEE303 pKa = 4.47CLLRR307 pKa = 11.84RR308 pKa = 11.84VSPAHH313 pKa = 5.38TALVIAYY320 pKa = 9.24DD321 pKa = 3.75GG322 pKa = 3.79
Molecular weight: 33.84 kDa
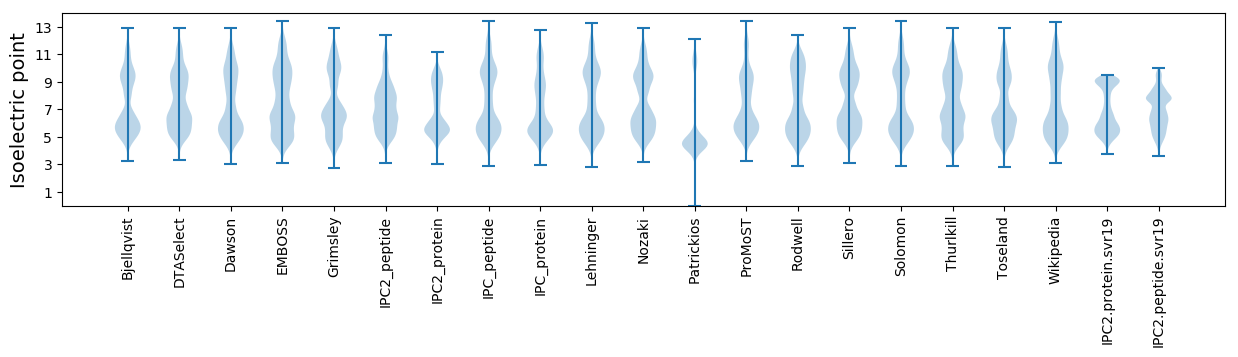
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2G8MDM3|A0A2G8MDM3_9RHIZ Potassium transporter OS=Methylobacterium frigidaeris OX=2038277 GN=CS379_23910 PE=4 SV=1
MM1 pKa = 7.66MIRR4 pKa = 11.84SNALRR9 pKa = 11.84AGLVLLALLAGLLTASPMVVTGIIAALGLAGAVALAGTRR48 pKa = 11.84AQQAPVLVPVRR59 pKa = 11.84ARR61 pKa = 11.84NRR63 pKa = 11.84RR64 pKa = 11.84RR65 pKa = 11.84RR66 pKa = 3.51
MM1 pKa = 7.66MIRR4 pKa = 11.84SNALRR9 pKa = 11.84AGLVLLALLAGLLTASPMVVTGIIAALGLAGAVALAGTRR48 pKa = 11.84AQQAPVLVPVRR59 pKa = 11.84ARR61 pKa = 11.84NRR63 pKa = 11.84RR64 pKa = 11.84RR65 pKa = 11.84RR66 pKa = 3.51
Molecular weight: 6.8 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1677552 |
20 |
2481 |
260.8 |
27.95 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.507 ± 0.052 | 0.81 ± 0.01 |
5.412 ± 0.027 | 5.524 ± 0.027 |
3.283 ± 0.02 | 9.239 ± 0.033 |
1.969 ± 0.014 | 4.204 ± 0.021 |
2.304 ± 0.03 | 10.671 ± 0.045 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.07 ± 0.015 | 1.946 ± 0.016 |
5.988 ± 0.029 | 2.746 ± 0.022 |
8.319 ± 0.031 | 4.695 ± 0.022 |
5.285 ± 0.022 | 7.773 ± 0.025 |
1.259 ± 0.012 | 1.993 ± 0.015 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
