
Broad bean mottle virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Alsuviricetes; Martellivirales; Bromoviridae; Bromovirus
Average proteome isoelectric point is 7.21
Get precalculated fractions of proteins
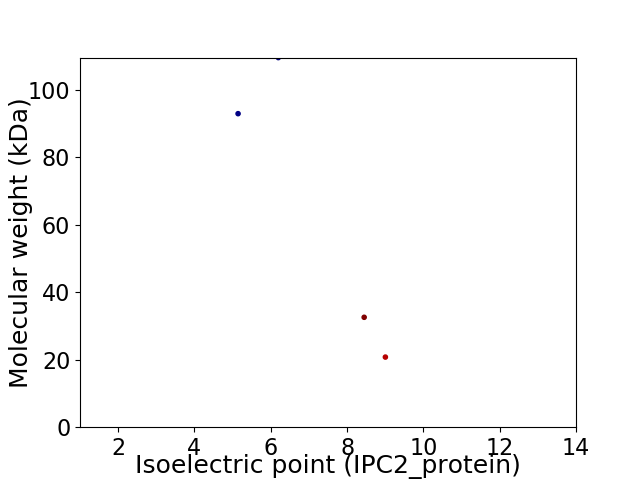
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
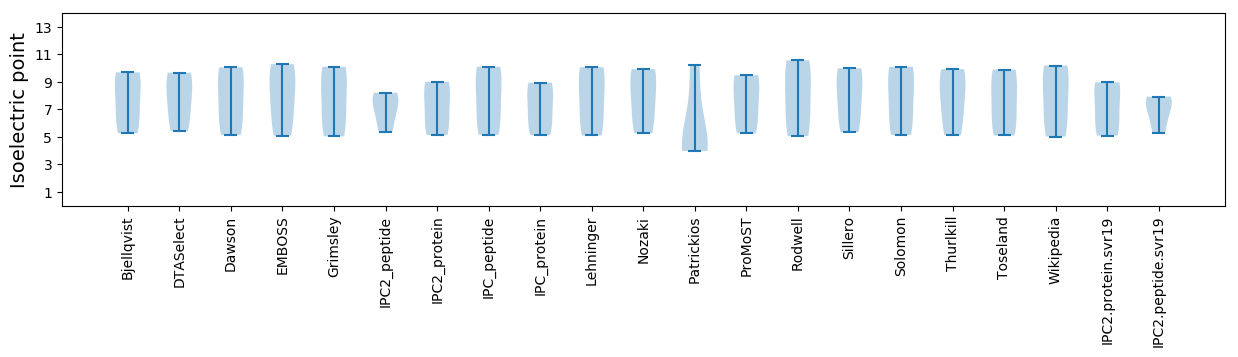
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|Q00020|1A_BBMV Replication protein 1a OS=Broad bean mottle virus OX=12301 GN=ORF1a PE=3 SV=1
MM1 pKa = 7.42SKK3 pKa = 10.08FVAADD8 pKa = 3.75EE9 pKa = 4.78YY10 pKa = 10.74IVPSFQWLLGPTSSQVEE27 pKa = 4.42SFDD30 pKa = 3.75SSVAGWIKK38 pKa = 10.35RR39 pKa = 11.84YY40 pKa = 8.89EE41 pKa = 4.38DD42 pKa = 3.49EE43 pKa = 4.45SKK45 pKa = 10.83RR46 pKa = 11.84PEE48 pKa = 4.01STVDD52 pKa = 3.66GSCEE56 pKa = 4.15SFVLAVKK63 pKa = 10.03PVMIGGQCEE72 pKa = 3.82PAYY75 pKa = 10.53DD76 pKa = 3.61QAKK79 pKa = 8.55WAEE82 pKa = 4.05TCLNVTNLASGLTGVRR98 pKa = 11.84LIPLPEE104 pKa = 4.23MARR107 pKa = 11.84MIYY110 pKa = 10.37LDD112 pKa = 4.29EE113 pKa = 4.82EE114 pKa = 4.61DD115 pKa = 4.31SFVDD119 pKa = 4.07EE120 pKa = 5.42SEE122 pKa = 4.09VDD124 pKa = 2.9DD125 pKa = 5.08WYY127 pKa = 10.55PEE129 pKa = 3.95DD130 pKa = 3.63TSDD133 pKa = 3.86GFEE136 pKa = 4.13YY137 pKa = 10.85LSADD141 pKa = 3.38GSDD144 pKa = 3.33YY145 pKa = 10.91HH146 pKa = 7.32QNSEE150 pKa = 4.25PEE152 pKa = 4.13EE153 pKa = 4.06EE154 pKa = 4.83VLDD157 pKa = 4.08KK158 pKa = 11.77SNGTLEE164 pKa = 4.26SEE166 pKa = 4.75EE167 pKa = 4.42ARR169 pKa = 11.84HH170 pKa = 4.91EE171 pKa = 4.16TDD173 pKa = 3.12VKK175 pKa = 10.67SICSFRR181 pKa = 11.84DD182 pKa = 3.2IPVEE186 pKa = 3.76TTMGHH191 pKa = 6.62RR192 pKa = 11.84YY193 pKa = 8.8LALSEE198 pKa = 4.21EE199 pKa = 4.45FASIEE204 pKa = 3.4VDD206 pKa = 3.66YY207 pKa = 10.8QVSSIVTPLNKK218 pKa = 10.14GSIYY222 pKa = 8.04MTLDD226 pKa = 3.1VHH228 pKa = 7.04EE229 pKa = 4.53EE230 pKa = 3.93THH232 pKa = 6.4PKK234 pKa = 10.52SGIQDD239 pKa = 3.33KK240 pKa = 11.27ASIEE244 pKa = 4.0RR245 pKa = 11.84LEE247 pKa = 4.38AAGHH251 pKa = 5.89NALKK255 pKa = 8.55THH257 pKa = 7.21AYY259 pKa = 10.22FDD261 pKa = 3.62DD262 pKa = 4.13SYY264 pKa = 11.85YY265 pKa = 11.02EE266 pKa = 4.16GFEE269 pKa = 3.96EE270 pKa = 5.05SADD273 pKa = 3.76FSSDD277 pKa = 3.39FQRR280 pKa = 11.84LKK282 pKa = 10.48IKK284 pKa = 10.23QSHH287 pKa = 5.11VDD289 pKa = 3.13WYY291 pKa = 11.03KK292 pKa = 11.4DD293 pKa = 3.11PDD295 pKa = 3.77KK296 pKa = 11.13FFEE299 pKa = 4.26PVLNFGASSRR309 pKa = 11.84RR310 pKa = 11.84VGSQKK315 pKa = 9.43TVLTALKK322 pKa = 9.92KK323 pKa = 10.5RR324 pKa = 11.84NADD327 pKa = 3.46VPEE330 pKa = 4.51LSDD333 pKa = 3.7VVDD336 pKa = 3.52ATEE339 pKa = 4.2VPICLRR345 pKa = 11.84QSYY348 pKa = 10.52LNIYY352 pKa = 9.6GADD355 pKa = 3.58CLFEE359 pKa = 4.58SFNIMAKK366 pKa = 10.43GLDD369 pKa = 3.35YY370 pKa = 10.82HH371 pKa = 7.07KK372 pKa = 10.13RR373 pKa = 11.84WKK375 pKa = 9.46SHH377 pKa = 6.68KK378 pKa = 8.6EE379 pKa = 3.86LQGVTLLCEE388 pKa = 4.34TNLQRR393 pKa = 11.84YY394 pKa = 6.05QHH396 pKa = 6.24MIKK399 pKa = 10.28SDD401 pKa = 3.62VKK403 pKa = 9.88PTVTDD408 pKa = 3.51TLHH411 pKa = 6.42VEE413 pKa = 4.03RR414 pKa = 11.84DD415 pKa = 3.51VPATITFHH423 pKa = 6.46GKK425 pKa = 10.12GVTSCFSPFFTACFEE440 pKa = 4.28KK441 pKa = 10.89FSLALRR447 pKa = 11.84EE448 pKa = 4.3SIYY451 pKa = 11.04VPIGKK456 pKa = 9.43ISSLEE461 pKa = 4.12LKK463 pKa = 10.7SKK465 pKa = 10.67ALNNKK470 pKa = 8.84FFLEE474 pKa = 4.34ADD476 pKa = 3.38LSKK479 pKa = 10.63FDD481 pKa = 3.7KK482 pKa = 11.18SQGEE486 pKa = 3.98LHH488 pKa = 7.1LEE490 pKa = 4.13FQRR493 pKa = 11.84LILINLGFPVPLTNWWCDD511 pKa = 3.78FHH513 pKa = 9.59RR514 pKa = 11.84SILSRR519 pKa = 11.84WTLRR523 pKa = 11.84PGVSIPLLSKK533 pKa = 8.57TNRR536 pKa = 11.84GCFHH540 pKa = 7.22LLWEE544 pKa = 4.7HH545 pKa = 6.57FSDD548 pKa = 3.82NAMMAYY554 pKa = 9.79CFDD557 pKa = 3.52MSTAEE562 pKa = 4.07LAMFSGDD569 pKa = 4.01DD570 pKa = 3.53SLVICGSKK578 pKa = 10.68PEE580 pKa = 4.08FDD582 pKa = 3.61PGVFQSLFNMEE593 pKa = 4.61VKK595 pKa = 10.87VMDD598 pKa = 4.34PSLPYY603 pKa = 9.9ICSKK607 pKa = 10.64FLLEE611 pKa = 4.83SEE613 pKa = 4.49FGDD616 pKa = 3.83VFSVPDD622 pKa = 3.89PMRR625 pKa = 11.84EE626 pKa = 3.6IQRR629 pKa = 11.84LQKK632 pKa = 10.53RR633 pKa = 11.84KK634 pKa = 9.4IPKK637 pKa = 9.49DD638 pKa = 3.73VQVLRR643 pKa = 11.84ADD645 pKa = 3.82FDD647 pKa = 4.49SFCDD651 pKa = 3.16RR652 pKa = 11.84MKK654 pKa = 10.89FLDD657 pKa = 3.93RR658 pKa = 11.84LSEE661 pKa = 3.96LSLSVLCRR669 pKa = 11.84LTALKK674 pKa = 10.18YY675 pKa = 9.51CKK677 pKa = 9.71PGIEE681 pKa = 4.06GDD683 pKa = 3.48VRR685 pKa = 11.84AWLSGFAYY693 pKa = 10.27YY694 pKa = 10.4RR695 pKa = 11.84EE696 pKa = 3.88NFLRR700 pKa = 11.84YY701 pKa = 8.99SEE703 pKa = 5.59CYY705 pKa = 8.11VTDD708 pKa = 5.3GIHH711 pKa = 6.97CYY713 pKa = 10.26RR714 pKa = 11.84RR715 pKa = 11.84VDD717 pKa = 3.15PMSRR721 pKa = 11.84FKK723 pKa = 10.88PIDD726 pKa = 3.11KK727 pKa = 9.97FQRR730 pKa = 11.84SDD732 pKa = 3.69KK733 pKa = 10.73EE734 pKa = 4.19WFHH737 pKa = 6.99DD738 pKa = 3.12WRR740 pKa = 11.84NNEE743 pKa = 4.08FPKK746 pKa = 10.9KK747 pKa = 10.12PIDD750 pKa = 3.18KK751 pKa = 8.41MARR754 pKa = 11.84MFGAYY759 pKa = 9.12KK760 pKa = 10.75GPVNNDD766 pKa = 2.63RR767 pKa = 11.84VEE769 pKa = 4.39RR770 pKa = 11.84KK771 pKa = 9.71AKK773 pKa = 10.09YY774 pKa = 9.23KK775 pKa = 10.93VNAAMHH781 pKa = 6.95DD782 pKa = 4.06PFALAYY788 pKa = 8.28EE789 pKa = 4.0RR790 pKa = 11.84RR791 pKa = 11.84YY792 pKa = 9.84VQEE795 pKa = 5.3LEE797 pKa = 4.27LKK799 pKa = 9.83NDD801 pKa = 3.6KK802 pKa = 10.83GNCSVLRR809 pKa = 11.84EE810 pKa = 4.12
MM1 pKa = 7.42SKK3 pKa = 10.08FVAADD8 pKa = 3.75EE9 pKa = 4.78YY10 pKa = 10.74IVPSFQWLLGPTSSQVEE27 pKa = 4.42SFDD30 pKa = 3.75SSVAGWIKK38 pKa = 10.35RR39 pKa = 11.84YY40 pKa = 8.89EE41 pKa = 4.38DD42 pKa = 3.49EE43 pKa = 4.45SKK45 pKa = 10.83RR46 pKa = 11.84PEE48 pKa = 4.01STVDD52 pKa = 3.66GSCEE56 pKa = 4.15SFVLAVKK63 pKa = 10.03PVMIGGQCEE72 pKa = 3.82PAYY75 pKa = 10.53DD76 pKa = 3.61QAKK79 pKa = 8.55WAEE82 pKa = 4.05TCLNVTNLASGLTGVRR98 pKa = 11.84LIPLPEE104 pKa = 4.23MARR107 pKa = 11.84MIYY110 pKa = 10.37LDD112 pKa = 4.29EE113 pKa = 4.82EE114 pKa = 4.61DD115 pKa = 4.31SFVDD119 pKa = 4.07EE120 pKa = 5.42SEE122 pKa = 4.09VDD124 pKa = 2.9DD125 pKa = 5.08WYY127 pKa = 10.55PEE129 pKa = 3.95DD130 pKa = 3.63TSDD133 pKa = 3.86GFEE136 pKa = 4.13YY137 pKa = 10.85LSADD141 pKa = 3.38GSDD144 pKa = 3.33YY145 pKa = 10.91HH146 pKa = 7.32QNSEE150 pKa = 4.25PEE152 pKa = 4.13EE153 pKa = 4.06EE154 pKa = 4.83VLDD157 pKa = 4.08KK158 pKa = 11.77SNGTLEE164 pKa = 4.26SEE166 pKa = 4.75EE167 pKa = 4.42ARR169 pKa = 11.84HH170 pKa = 4.91EE171 pKa = 4.16TDD173 pKa = 3.12VKK175 pKa = 10.67SICSFRR181 pKa = 11.84DD182 pKa = 3.2IPVEE186 pKa = 3.76TTMGHH191 pKa = 6.62RR192 pKa = 11.84YY193 pKa = 8.8LALSEE198 pKa = 4.21EE199 pKa = 4.45FASIEE204 pKa = 3.4VDD206 pKa = 3.66YY207 pKa = 10.8QVSSIVTPLNKK218 pKa = 10.14GSIYY222 pKa = 8.04MTLDD226 pKa = 3.1VHH228 pKa = 7.04EE229 pKa = 4.53EE230 pKa = 3.93THH232 pKa = 6.4PKK234 pKa = 10.52SGIQDD239 pKa = 3.33KK240 pKa = 11.27ASIEE244 pKa = 4.0RR245 pKa = 11.84LEE247 pKa = 4.38AAGHH251 pKa = 5.89NALKK255 pKa = 8.55THH257 pKa = 7.21AYY259 pKa = 10.22FDD261 pKa = 3.62DD262 pKa = 4.13SYY264 pKa = 11.85YY265 pKa = 11.02EE266 pKa = 4.16GFEE269 pKa = 3.96EE270 pKa = 5.05SADD273 pKa = 3.76FSSDD277 pKa = 3.39FQRR280 pKa = 11.84LKK282 pKa = 10.48IKK284 pKa = 10.23QSHH287 pKa = 5.11VDD289 pKa = 3.13WYY291 pKa = 11.03KK292 pKa = 11.4DD293 pKa = 3.11PDD295 pKa = 3.77KK296 pKa = 11.13FFEE299 pKa = 4.26PVLNFGASSRR309 pKa = 11.84RR310 pKa = 11.84VGSQKK315 pKa = 9.43TVLTALKK322 pKa = 9.92KK323 pKa = 10.5RR324 pKa = 11.84NADD327 pKa = 3.46VPEE330 pKa = 4.51LSDD333 pKa = 3.7VVDD336 pKa = 3.52ATEE339 pKa = 4.2VPICLRR345 pKa = 11.84QSYY348 pKa = 10.52LNIYY352 pKa = 9.6GADD355 pKa = 3.58CLFEE359 pKa = 4.58SFNIMAKK366 pKa = 10.43GLDD369 pKa = 3.35YY370 pKa = 10.82HH371 pKa = 7.07KK372 pKa = 10.13RR373 pKa = 11.84WKK375 pKa = 9.46SHH377 pKa = 6.68KK378 pKa = 8.6EE379 pKa = 3.86LQGVTLLCEE388 pKa = 4.34TNLQRR393 pKa = 11.84YY394 pKa = 6.05QHH396 pKa = 6.24MIKK399 pKa = 10.28SDD401 pKa = 3.62VKK403 pKa = 9.88PTVTDD408 pKa = 3.51TLHH411 pKa = 6.42VEE413 pKa = 4.03RR414 pKa = 11.84DD415 pKa = 3.51VPATITFHH423 pKa = 6.46GKK425 pKa = 10.12GVTSCFSPFFTACFEE440 pKa = 4.28KK441 pKa = 10.89FSLALRR447 pKa = 11.84EE448 pKa = 4.3SIYY451 pKa = 11.04VPIGKK456 pKa = 9.43ISSLEE461 pKa = 4.12LKK463 pKa = 10.7SKK465 pKa = 10.67ALNNKK470 pKa = 8.84FFLEE474 pKa = 4.34ADD476 pKa = 3.38LSKK479 pKa = 10.63FDD481 pKa = 3.7KK482 pKa = 11.18SQGEE486 pKa = 3.98LHH488 pKa = 7.1LEE490 pKa = 4.13FQRR493 pKa = 11.84LILINLGFPVPLTNWWCDD511 pKa = 3.78FHH513 pKa = 9.59RR514 pKa = 11.84SILSRR519 pKa = 11.84WTLRR523 pKa = 11.84PGVSIPLLSKK533 pKa = 8.57TNRR536 pKa = 11.84GCFHH540 pKa = 7.22LLWEE544 pKa = 4.7HH545 pKa = 6.57FSDD548 pKa = 3.82NAMMAYY554 pKa = 9.79CFDD557 pKa = 3.52MSTAEE562 pKa = 4.07LAMFSGDD569 pKa = 4.01DD570 pKa = 3.53SLVICGSKK578 pKa = 10.68PEE580 pKa = 4.08FDD582 pKa = 3.61PGVFQSLFNMEE593 pKa = 4.61VKK595 pKa = 10.87VMDD598 pKa = 4.34PSLPYY603 pKa = 9.9ICSKK607 pKa = 10.64FLLEE611 pKa = 4.83SEE613 pKa = 4.49FGDD616 pKa = 3.83VFSVPDD622 pKa = 3.89PMRR625 pKa = 11.84EE626 pKa = 3.6IQRR629 pKa = 11.84LQKK632 pKa = 10.53RR633 pKa = 11.84KK634 pKa = 9.4IPKK637 pKa = 9.49DD638 pKa = 3.73VQVLRR643 pKa = 11.84ADD645 pKa = 3.82FDD647 pKa = 4.49SFCDD651 pKa = 3.16RR652 pKa = 11.84MKK654 pKa = 10.89FLDD657 pKa = 3.93RR658 pKa = 11.84LSEE661 pKa = 3.96LSLSVLCRR669 pKa = 11.84LTALKK674 pKa = 10.18YY675 pKa = 9.51CKK677 pKa = 9.71PGIEE681 pKa = 4.06GDD683 pKa = 3.48VRR685 pKa = 11.84AWLSGFAYY693 pKa = 10.27YY694 pKa = 10.4RR695 pKa = 11.84EE696 pKa = 3.88NFLRR700 pKa = 11.84YY701 pKa = 8.99SEE703 pKa = 5.59CYY705 pKa = 8.11VTDD708 pKa = 5.3GIHH711 pKa = 6.97CYY713 pKa = 10.26RR714 pKa = 11.84RR715 pKa = 11.84VDD717 pKa = 3.15PMSRR721 pKa = 11.84FKK723 pKa = 10.88PIDD726 pKa = 3.11KK727 pKa = 9.97FQRR730 pKa = 11.84SDD732 pKa = 3.69KK733 pKa = 10.73EE734 pKa = 4.19WFHH737 pKa = 6.99DD738 pKa = 3.12WRR740 pKa = 11.84NNEE743 pKa = 4.08FPKK746 pKa = 10.9KK747 pKa = 10.12PIDD750 pKa = 3.18KK751 pKa = 8.41MARR754 pKa = 11.84MFGAYY759 pKa = 9.12KK760 pKa = 10.75GPVNNDD766 pKa = 2.63RR767 pKa = 11.84VEE769 pKa = 4.39RR770 pKa = 11.84KK771 pKa = 9.71AKK773 pKa = 10.09YY774 pKa = 9.23KK775 pKa = 10.93VNAAMHH781 pKa = 6.95DD782 pKa = 4.06PFALAYY788 pKa = 8.28EE789 pKa = 4.0RR790 pKa = 11.84RR791 pKa = 11.84YY792 pKa = 9.84VQEE795 pKa = 5.3LEE797 pKa = 4.27LKK799 pKa = 9.83NDD801 pKa = 3.6KK802 pKa = 10.83GNCSVLRR809 pKa = 11.84EE810 pKa = 4.12
Molecular weight: 92.99 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|P24403|MVP_BBMV Movement protein OS=Broad bean mottle virus OX=12301 GN=ORF3a PE=3 SV=1
MM1 pKa = 6.49TTSATGKK8 pKa = 10.21ALNRR12 pKa = 11.84KK13 pKa = 7.38QRR15 pKa = 11.84RR16 pKa = 11.84ALNRR20 pKa = 11.84SNRR23 pKa = 11.84LRR25 pKa = 11.84KK26 pKa = 9.06EE27 pKa = 3.81FQPVIVEE34 pKa = 4.17PLASGQAVSLKK45 pKa = 9.65TRR47 pKa = 11.84TGYY50 pKa = 10.71CVTQFVSNNPEE61 pKa = 3.72VKK63 pKa = 10.05AKK65 pKa = 10.16EE66 pKa = 4.17VVSVSVKK73 pKa = 10.77LPDD76 pKa = 3.73HH77 pKa = 6.46LAVEE81 pKa = 4.77ANRR84 pKa = 11.84ALKK87 pKa = 10.29VGRR90 pKa = 11.84ISILLGLLPTVAGTVKK106 pKa = 10.32VCLTEE111 pKa = 4.7KK112 pKa = 10.21QDD114 pKa = 3.88SPAEE118 pKa = 4.14SFKK121 pKa = 10.95RR122 pKa = 11.84ALAVADD128 pKa = 3.63SSKK131 pKa = 10.78EE132 pKa = 3.91VASAFYY138 pKa = 11.03VDD140 pKa = 3.76GFKK143 pKa = 10.89DD144 pKa = 3.54VSLGDD149 pKa = 3.83LEE151 pKa = 5.63KK152 pKa = 11.13DD153 pKa = 3.28LSIYY157 pKa = 10.34LYY159 pKa = 10.96SEE161 pKa = 4.05AALAANSIRR170 pKa = 11.84IRR172 pKa = 11.84MEE174 pKa = 3.69VEE176 pKa = 3.42HH177 pKa = 6.44VMPKK181 pKa = 10.27FITRR185 pKa = 11.84FSPFAA190 pKa = 5.09
MM1 pKa = 6.49TTSATGKK8 pKa = 10.21ALNRR12 pKa = 11.84KK13 pKa = 7.38QRR15 pKa = 11.84RR16 pKa = 11.84ALNRR20 pKa = 11.84SNRR23 pKa = 11.84LRR25 pKa = 11.84KK26 pKa = 9.06EE27 pKa = 3.81FQPVIVEE34 pKa = 4.17PLASGQAVSLKK45 pKa = 9.65TRR47 pKa = 11.84TGYY50 pKa = 10.71CVTQFVSNNPEE61 pKa = 3.72VKK63 pKa = 10.05AKK65 pKa = 10.16EE66 pKa = 4.17VVSVSVKK73 pKa = 10.77LPDD76 pKa = 3.73HH77 pKa = 6.46LAVEE81 pKa = 4.77ANRR84 pKa = 11.84ALKK87 pKa = 10.29VGRR90 pKa = 11.84ISILLGLLPTVAGTVKK106 pKa = 10.32VCLTEE111 pKa = 4.7KK112 pKa = 10.21QDD114 pKa = 3.88SPAEE118 pKa = 4.14SFKK121 pKa = 10.95RR122 pKa = 11.84ALAVADD128 pKa = 3.63SSKK131 pKa = 10.78EE132 pKa = 3.91VASAFYY138 pKa = 11.03VDD140 pKa = 3.76GFKK143 pKa = 10.89DD144 pKa = 3.54VSLGDD149 pKa = 3.83LEE151 pKa = 5.63KK152 pKa = 11.13DD153 pKa = 3.28LSIYY157 pKa = 10.34LYY159 pKa = 10.96SEE161 pKa = 4.05AALAANSIRR170 pKa = 11.84IRR172 pKa = 11.84MEE174 pKa = 3.69VEE176 pKa = 3.42HH177 pKa = 6.44VMPKK181 pKa = 10.27FITRR185 pKa = 11.84FSPFAA190 pKa = 5.09
Molecular weight: 20.8 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2261 |
190 |
966 |
565.3 |
64.0 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.678 ± 0.752 | 2.565 ± 0.462 |
6.236 ± 0.755 | 7.298 ± 0.525 |
4.865 ± 0.606 | 5.263 ± 0.373 |
2.433 ± 0.322 | 4.777 ± 0.384 |
6.855 ± 0.178 | 8.536 ± 0.371 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.477 ± 0.218 | 4.069 ± 0.404 |
3.671 ± 0.695 | 2.521 ± 0.094 |
5.927 ± 0.291 | 8.89 ± 0.727 |
4.909 ± 0.432 | 7.386 ± 0.612 |
1.327 ± 0.394 | 3.317 ± 0.289 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
