
Lake Victoria marburgvirus (strain Musoke-80) (MARV) (Marburg virus (strain Kenya/Musoke/1980))
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Filoviridae; Marburgvirus; Marburg marburgvirus
Average proteome isoelectric point is 7.22
Get precalculated fractions of proteins
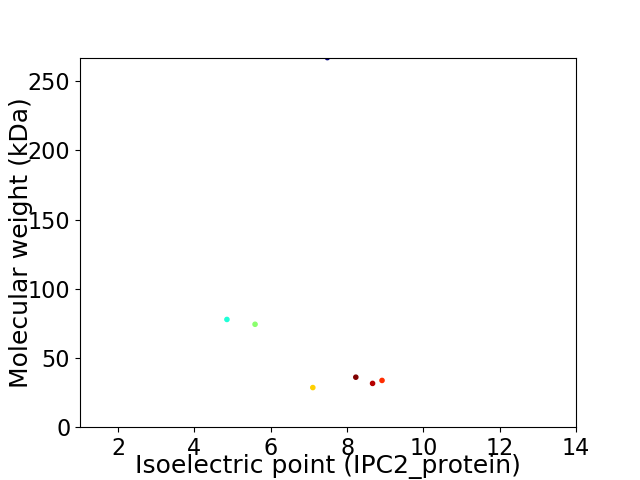
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
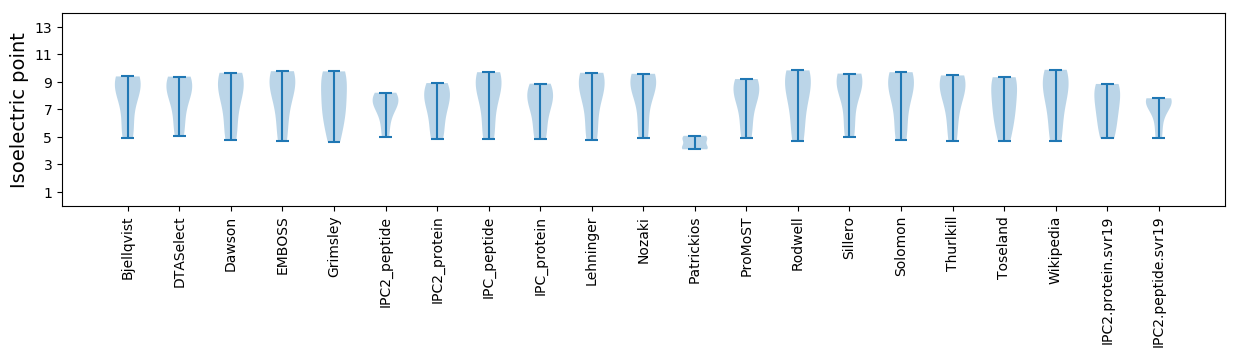
* You can choose from 21 different methods for calculating isoelectric point
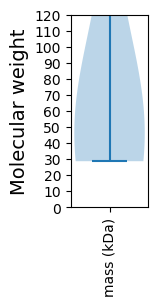
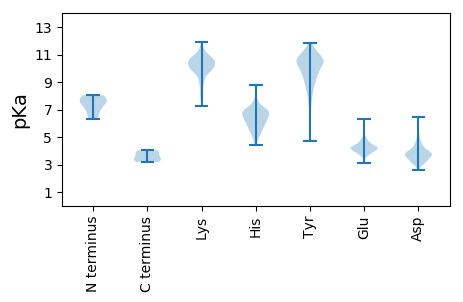
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|P31352|L_MABVM RNA-directed RNA polymerase L OS=Lake Victoria marburgvirus (strain Musoke-80) OX=33727 GN=L PE=3 SV=2
MM1 pKa = 8.04DD2 pKa = 4.52LHH4 pKa = 7.48SLLEE8 pKa = 4.65LGTKK12 pKa = 7.93PTAPHH17 pKa = 5.24VRR19 pKa = 11.84NKK21 pKa = 10.41KK22 pKa = 10.42VILFDD27 pKa = 3.75TNHH30 pKa = 5.59QVSICNQIIDD40 pKa = 5.28AINSGIDD47 pKa = 3.76LGDD50 pKa = 3.75LLEE53 pKa = 5.28GGLLTLCVEE62 pKa = 4.95HH63 pKa = 6.82YY64 pKa = 10.8YY65 pKa = 11.25NSDD68 pKa = 3.0KK69 pKa = 11.56DD70 pKa = 3.74KK71 pKa = 11.39FNTSPVAKK79 pKa = 9.16YY80 pKa = 10.57LRR82 pKa = 11.84DD83 pKa = 3.24AGYY86 pKa = 10.74EE87 pKa = 3.69FDD89 pKa = 4.23VIKK92 pKa = 11.05NADD95 pKa = 3.2ATRR98 pKa = 11.84FLDD101 pKa = 3.53VSPNEE106 pKa = 3.86PHH108 pKa = 6.52YY109 pKa = 11.36SPLILALKK117 pKa = 8.55TLEE120 pKa = 4.17STEE123 pKa = 4.17SQRR126 pKa = 11.84GRR128 pKa = 11.84IGLFLSFCSLFLPKK142 pKa = 10.41LVVGDD147 pKa = 3.54RR148 pKa = 11.84ASIEE152 pKa = 3.82KK153 pKa = 10.41ALRR156 pKa = 11.84QVTVHH161 pKa = 5.56QEE163 pKa = 3.93QGIVTYY169 pKa = 9.9PNHH172 pKa = 6.71WLTTGHH178 pKa = 6.17MKK180 pKa = 10.61VIFGILRR187 pKa = 11.84SSFILKK193 pKa = 9.98FVLIHH198 pKa = 5.96QGVNLVTGHH207 pKa = 7.24DD208 pKa = 4.17AYY210 pKa = 11.14DD211 pKa = 4.07SIISNSVGQTRR222 pKa = 11.84FSGLLIVKK230 pKa = 8.21TVLEE234 pKa = 5.06FILQKK239 pKa = 9.7TDD241 pKa = 2.82SGVTLHH247 pKa = 6.57PLVRR251 pKa = 11.84TSKK254 pKa = 10.47VKK256 pKa = 11.14NEE258 pKa = 3.81VASFKK263 pKa = 10.58QALSNLARR271 pKa = 11.84HH272 pKa = 6.11GEE274 pKa = 4.08YY275 pKa = 10.89APFARR280 pKa = 11.84VLNLSGINNLEE291 pKa = 4.3HH292 pKa = 6.95GLYY295 pKa = 9.39PQLSAIALGVATAHH309 pKa = 6.58GSTLAGVNVGEE320 pKa = 4.64QYY322 pKa = 10.17QQLRR326 pKa = 11.84EE327 pKa = 4.07AAHH330 pKa = 6.03DD331 pKa = 4.04AEE333 pKa = 4.82VKK335 pKa = 9.37LQRR338 pKa = 11.84RR339 pKa = 11.84HH340 pKa = 4.63EE341 pKa = 4.21HH342 pKa = 6.06QEE344 pKa = 3.48IQAIAEE350 pKa = 4.16DD351 pKa = 3.91DD352 pKa = 3.91EE353 pKa = 4.29EE354 pKa = 5.24RR355 pKa = 11.84KK356 pKa = 9.94ILEE359 pKa = 4.01QFHH362 pKa = 5.72LQKK365 pKa = 9.98TEE367 pKa = 3.73ITHH370 pKa = 5.44SQTLAVLSQKK380 pKa = 10.21RR381 pKa = 11.84EE382 pKa = 3.81KK383 pKa = 10.42LARR386 pKa = 11.84LAAEE390 pKa = 3.79IEE392 pKa = 4.2NNIVEE397 pKa = 4.38DD398 pKa = 3.53QGFKK402 pKa = 10.11QSQNRR407 pKa = 11.84VSQSFLNDD415 pKa = 3.43PTPVEE420 pKa = 4.29VTVQARR426 pKa = 11.84PMNRR430 pKa = 11.84PTALPPPVDD439 pKa = 4.08DD440 pKa = 5.38KK441 pKa = 11.53IEE443 pKa = 4.31HH444 pKa = 6.83EE445 pKa = 4.33STEE448 pKa = 4.41DD449 pKa = 3.42SSSSSSFVDD458 pKa = 3.95LNDD461 pKa = 3.61PFALLNEE468 pKa = 4.95DD469 pKa = 4.71EE470 pKa = 4.81DD471 pKa = 4.44TLDD474 pKa = 4.23DD475 pKa = 3.78SVMIPGTTSRR485 pKa = 11.84EE486 pKa = 3.9FQGIPEE492 pKa = 4.81PPRR495 pKa = 11.84QSQDD499 pKa = 3.14LNNSQGKK506 pKa = 9.17QEE508 pKa = 4.49DD509 pKa = 4.06EE510 pKa = 3.96STNRR514 pKa = 11.84IKK516 pKa = 10.84KK517 pKa = 9.59QFLRR521 pKa = 11.84YY522 pKa = 9.42QEE524 pKa = 4.57LPPVQEE530 pKa = 4.9DD531 pKa = 4.01DD532 pKa = 3.58EE533 pKa = 5.27SEE535 pKa = 4.33YY536 pKa = 8.54TTDD539 pKa = 3.24SQEE542 pKa = 5.59SIDD545 pKa = 4.96QPGSDD550 pKa = 3.86NEE552 pKa = 4.3QGVDD556 pKa = 3.96LPPPPLYY563 pKa = 10.67AQEE566 pKa = 4.25KK567 pKa = 8.18RR568 pKa = 11.84QDD570 pKa = 4.51PIQHH574 pKa = 6.82PAANPQDD581 pKa = 3.85PFGSIGDD588 pKa = 3.74VNGDD592 pKa = 3.0ILEE595 pKa = 5.1PIRR598 pKa = 11.84SPSSPSAPQEE608 pKa = 3.81DD609 pKa = 3.79TRR611 pKa = 11.84MRR613 pKa = 11.84EE614 pKa = 4.22AYY616 pKa = 9.32EE617 pKa = 4.06LSPDD621 pKa = 3.66FTNDD625 pKa = 3.84EE626 pKa = 4.8DD627 pKa = 4.68NQQNWPQRR635 pKa = 11.84VVTKK639 pKa = 10.47KK640 pKa = 10.64GRR642 pKa = 11.84TFLYY646 pKa = 10.4PNDD649 pKa = 4.25LLQTNPPEE657 pKa = 4.24SLITALVEE665 pKa = 4.45EE666 pKa = 4.58YY667 pKa = 10.69QNPVSAKK674 pKa = 9.4EE675 pKa = 4.17LQADD679 pKa = 3.88WPDD682 pKa = 3.06MSFDD686 pKa = 3.66EE687 pKa = 4.47RR688 pKa = 11.84RR689 pKa = 11.84HH690 pKa = 4.6VAMNLL695 pKa = 3.25
MM1 pKa = 8.04DD2 pKa = 4.52LHH4 pKa = 7.48SLLEE8 pKa = 4.65LGTKK12 pKa = 7.93PTAPHH17 pKa = 5.24VRR19 pKa = 11.84NKK21 pKa = 10.41KK22 pKa = 10.42VILFDD27 pKa = 3.75TNHH30 pKa = 5.59QVSICNQIIDD40 pKa = 5.28AINSGIDD47 pKa = 3.76LGDD50 pKa = 3.75LLEE53 pKa = 5.28GGLLTLCVEE62 pKa = 4.95HH63 pKa = 6.82YY64 pKa = 10.8YY65 pKa = 11.25NSDD68 pKa = 3.0KK69 pKa = 11.56DD70 pKa = 3.74KK71 pKa = 11.39FNTSPVAKK79 pKa = 9.16YY80 pKa = 10.57LRR82 pKa = 11.84DD83 pKa = 3.24AGYY86 pKa = 10.74EE87 pKa = 3.69FDD89 pKa = 4.23VIKK92 pKa = 11.05NADD95 pKa = 3.2ATRR98 pKa = 11.84FLDD101 pKa = 3.53VSPNEE106 pKa = 3.86PHH108 pKa = 6.52YY109 pKa = 11.36SPLILALKK117 pKa = 8.55TLEE120 pKa = 4.17STEE123 pKa = 4.17SQRR126 pKa = 11.84GRR128 pKa = 11.84IGLFLSFCSLFLPKK142 pKa = 10.41LVVGDD147 pKa = 3.54RR148 pKa = 11.84ASIEE152 pKa = 3.82KK153 pKa = 10.41ALRR156 pKa = 11.84QVTVHH161 pKa = 5.56QEE163 pKa = 3.93QGIVTYY169 pKa = 9.9PNHH172 pKa = 6.71WLTTGHH178 pKa = 6.17MKK180 pKa = 10.61VIFGILRR187 pKa = 11.84SSFILKK193 pKa = 9.98FVLIHH198 pKa = 5.96QGVNLVTGHH207 pKa = 7.24DD208 pKa = 4.17AYY210 pKa = 11.14DD211 pKa = 4.07SIISNSVGQTRR222 pKa = 11.84FSGLLIVKK230 pKa = 8.21TVLEE234 pKa = 5.06FILQKK239 pKa = 9.7TDD241 pKa = 2.82SGVTLHH247 pKa = 6.57PLVRR251 pKa = 11.84TSKK254 pKa = 10.47VKK256 pKa = 11.14NEE258 pKa = 3.81VASFKK263 pKa = 10.58QALSNLARR271 pKa = 11.84HH272 pKa = 6.11GEE274 pKa = 4.08YY275 pKa = 10.89APFARR280 pKa = 11.84VLNLSGINNLEE291 pKa = 4.3HH292 pKa = 6.95GLYY295 pKa = 9.39PQLSAIALGVATAHH309 pKa = 6.58GSTLAGVNVGEE320 pKa = 4.64QYY322 pKa = 10.17QQLRR326 pKa = 11.84EE327 pKa = 4.07AAHH330 pKa = 6.03DD331 pKa = 4.04AEE333 pKa = 4.82VKK335 pKa = 9.37LQRR338 pKa = 11.84RR339 pKa = 11.84HH340 pKa = 4.63EE341 pKa = 4.21HH342 pKa = 6.06QEE344 pKa = 3.48IQAIAEE350 pKa = 4.16DD351 pKa = 3.91DD352 pKa = 3.91EE353 pKa = 4.29EE354 pKa = 5.24RR355 pKa = 11.84KK356 pKa = 9.94ILEE359 pKa = 4.01QFHH362 pKa = 5.72LQKK365 pKa = 9.98TEE367 pKa = 3.73ITHH370 pKa = 5.44SQTLAVLSQKK380 pKa = 10.21RR381 pKa = 11.84EE382 pKa = 3.81KK383 pKa = 10.42LARR386 pKa = 11.84LAAEE390 pKa = 3.79IEE392 pKa = 4.2NNIVEE397 pKa = 4.38DD398 pKa = 3.53QGFKK402 pKa = 10.11QSQNRR407 pKa = 11.84VSQSFLNDD415 pKa = 3.43PTPVEE420 pKa = 4.29VTVQARR426 pKa = 11.84PMNRR430 pKa = 11.84PTALPPPVDD439 pKa = 4.08DD440 pKa = 5.38KK441 pKa = 11.53IEE443 pKa = 4.31HH444 pKa = 6.83EE445 pKa = 4.33STEE448 pKa = 4.41DD449 pKa = 3.42SSSSSSFVDD458 pKa = 3.95LNDD461 pKa = 3.61PFALLNEE468 pKa = 4.95DD469 pKa = 4.71EE470 pKa = 4.81DD471 pKa = 4.44TLDD474 pKa = 4.23DD475 pKa = 3.78SVMIPGTTSRR485 pKa = 11.84EE486 pKa = 3.9FQGIPEE492 pKa = 4.81PPRR495 pKa = 11.84QSQDD499 pKa = 3.14LNNSQGKK506 pKa = 9.17QEE508 pKa = 4.49DD509 pKa = 4.06EE510 pKa = 3.96STNRR514 pKa = 11.84IKK516 pKa = 10.84KK517 pKa = 9.59QFLRR521 pKa = 11.84YY522 pKa = 9.42QEE524 pKa = 4.57LPPVQEE530 pKa = 4.9DD531 pKa = 4.01DD532 pKa = 3.58EE533 pKa = 5.27SEE535 pKa = 4.33YY536 pKa = 8.54TTDD539 pKa = 3.24SQEE542 pKa = 5.59SIDD545 pKa = 4.96QPGSDD550 pKa = 3.86NEE552 pKa = 4.3QGVDD556 pKa = 3.96LPPPPLYY563 pKa = 10.67AQEE566 pKa = 4.25KK567 pKa = 8.18RR568 pKa = 11.84QDD570 pKa = 4.51PIQHH574 pKa = 6.82PAANPQDD581 pKa = 3.85PFGSIGDD588 pKa = 3.74VNGDD592 pKa = 3.0ILEE595 pKa = 5.1PIRR598 pKa = 11.84SPSSPSAPQEE608 pKa = 3.81DD609 pKa = 3.79TRR611 pKa = 11.84MRR613 pKa = 11.84EE614 pKa = 4.22AYY616 pKa = 9.32EE617 pKa = 4.06LSPDD621 pKa = 3.66FTNDD625 pKa = 3.84EE626 pKa = 4.8DD627 pKa = 4.68NQQNWPQRR635 pKa = 11.84VVTKK639 pKa = 10.47KK640 pKa = 10.64GRR642 pKa = 11.84TFLYY646 pKa = 10.4PNDD649 pKa = 4.25LLQTNPPEE657 pKa = 4.24SLITALVEE665 pKa = 4.45EE666 pKa = 4.58YY667 pKa = 10.69QNPVSAKK674 pKa = 9.4EE675 pKa = 4.17LQADD679 pKa = 3.88WPDD682 pKa = 3.06MSFDD686 pKa = 3.66EE687 pKa = 4.47RR688 pKa = 11.84RR689 pKa = 11.84HH690 pKa = 4.6VAMNLL695 pKa = 3.25
Molecular weight: 77.86 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|P35259|VP35_MABVM Polymerase cofactor VP35 OS=Lake Victoria marburgvirus (strain Musoke-80) OX=33727 GN=VP35 PE=1 SV=1
MM1 pKa = 6.29QQPRR5 pKa = 11.84GRR7 pKa = 11.84SRR9 pKa = 11.84TRR11 pKa = 11.84NHH13 pKa = 5.53QVTPTIYY20 pKa = 10.52HH21 pKa = 5.65EE22 pKa = 4.38TQLPSKK28 pKa = 9.74PHH30 pKa = 4.43YY31 pKa = 9.3TNYY34 pKa = 10.04HH35 pKa = 5.0PRR37 pKa = 11.84ARR39 pKa = 11.84SMSSTRR45 pKa = 11.84SSAEE49 pKa = 3.7SSPTNHH55 pKa = 6.78IPRR58 pKa = 11.84ARR60 pKa = 11.84PPSTFNLSKK69 pKa = 10.53PPPPPKK75 pKa = 10.56DD76 pKa = 2.85MCRR79 pKa = 11.84NMKK82 pKa = 10.06IGLPCADD89 pKa = 3.59PTCNRR94 pKa = 11.84DD95 pKa = 3.04HH96 pKa = 7.73DD97 pKa = 5.04LDD99 pKa = 3.86NLTNRR104 pKa = 11.84EE105 pKa = 4.3LLLLMARR112 pKa = 11.84KK113 pKa = 8.27MLPNTDD119 pKa = 2.85KK120 pKa = 10.91TFRR123 pKa = 11.84SPQDD127 pKa = 3.35CGSPSLSKK135 pKa = 10.84GLSKK139 pKa = 10.9DD140 pKa = 3.3KK141 pKa = 11.08QEE143 pKa = 3.93QTKK146 pKa = 10.92DD147 pKa = 3.38VLTLEE152 pKa = 4.31NLGHH156 pKa = 6.39ILSYY160 pKa = 9.29LHH162 pKa = 6.8RR163 pKa = 11.84SEE165 pKa = 4.91IGKK168 pKa = 9.78LDD170 pKa = 3.73EE171 pKa = 4.47TSLRR175 pKa = 11.84AALSLTCAGIRR186 pKa = 11.84KK187 pKa = 7.28TNRR190 pKa = 11.84SLINTMTEE198 pKa = 3.11LHH200 pKa = 6.53MNHH203 pKa = 6.48EE204 pKa = 4.89NLPQDD209 pKa = 3.27QNGVIKK215 pKa = 9.13QTYY218 pKa = 8.31TGIHH222 pKa = 6.7LDD224 pKa = 3.15KK225 pKa = 11.17GGQFEE230 pKa = 4.05AALWQGWDD238 pKa = 3.44KK239 pKa = 11.29RR240 pKa = 11.84SISLFVQAALYY251 pKa = 10.21VMNNIPCEE259 pKa = 4.11SSISVQASYY268 pKa = 11.72DD269 pKa = 3.75HH270 pKa = 7.22FILPQSQGKK279 pKa = 8.18GQQ281 pKa = 3.19
MM1 pKa = 6.29QQPRR5 pKa = 11.84GRR7 pKa = 11.84SRR9 pKa = 11.84TRR11 pKa = 11.84NHH13 pKa = 5.53QVTPTIYY20 pKa = 10.52HH21 pKa = 5.65EE22 pKa = 4.38TQLPSKK28 pKa = 9.74PHH30 pKa = 4.43YY31 pKa = 9.3TNYY34 pKa = 10.04HH35 pKa = 5.0PRR37 pKa = 11.84ARR39 pKa = 11.84SMSSTRR45 pKa = 11.84SSAEE49 pKa = 3.7SSPTNHH55 pKa = 6.78IPRR58 pKa = 11.84ARR60 pKa = 11.84PPSTFNLSKK69 pKa = 10.53PPPPPKK75 pKa = 10.56DD76 pKa = 2.85MCRR79 pKa = 11.84NMKK82 pKa = 10.06IGLPCADD89 pKa = 3.59PTCNRR94 pKa = 11.84DD95 pKa = 3.04HH96 pKa = 7.73DD97 pKa = 5.04LDD99 pKa = 3.86NLTNRR104 pKa = 11.84EE105 pKa = 4.3LLLLMARR112 pKa = 11.84KK113 pKa = 8.27MLPNTDD119 pKa = 2.85KK120 pKa = 10.91TFRR123 pKa = 11.84SPQDD127 pKa = 3.35CGSPSLSKK135 pKa = 10.84GLSKK139 pKa = 10.9DD140 pKa = 3.3KK141 pKa = 11.08QEE143 pKa = 3.93QTKK146 pKa = 10.92DD147 pKa = 3.38VLTLEE152 pKa = 4.31NLGHH156 pKa = 6.39ILSYY160 pKa = 9.29LHH162 pKa = 6.8RR163 pKa = 11.84SEE165 pKa = 4.91IGKK168 pKa = 9.78LDD170 pKa = 3.73EE171 pKa = 4.47TSLRR175 pKa = 11.84AALSLTCAGIRR186 pKa = 11.84KK187 pKa = 7.28TNRR190 pKa = 11.84SLINTMTEE198 pKa = 3.11LHH200 pKa = 6.53MNHH203 pKa = 6.48EE204 pKa = 4.89NLPQDD209 pKa = 3.27QNGVIKK215 pKa = 9.13QTYY218 pKa = 8.31TGIHH222 pKa = 6.7LDD224 pKa = 3.15KK225 pKa = 11.17GGQFEE230 pKa = 4.05AALWQGWDD238 pKa = 3.44KK239 pKa = 11.29RR240 pKa = 11.84SISLFVQAALYY251 pKa = 10.21VMNNIPCEE259 pKa = 4.11SSISVQASYY268 pKa = 11.72DD269 pKa = 3.75HH270 pKa = 7.22FILPQSQGKK279 pKa = 8.18GQQ281 pKa = 3.19
Molecular weight: 31.65 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4873 |
253 |
2331 |
696.1 |
78.51 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.459 ± 0.444 | 1.642 ± 0.364 |
5.007 ± 0.333 | 5.315 ± 0.41 |
4.207 ± 0.477 | 4.946 ± 0.413 |
2.873 ± 0.153 | 6.382 ± 0.389 |
5.52 ± 0.269 | 11.081 ± 0.69 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.826 ± 0.295 | 5.91 ± 0.462 |
5.582 ± 0.777 | 4.925 ± 0.391 |
4.638 ± 0.314 | 8.742 ± 0.497 |
6.628 ± 0.951 | 4.925 ± 0.604 |
1.231 ± 0.177 | 3.16 ± 0.586 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
