
Tai Forest reovirus
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Resentoviricetes; Reovirales; Reoviridae; Spinareovirinae; Coltivirus; Tai Forest coltivirus
Average proteome isoelectric point is 6.02
Get precalculated fractions of proteins
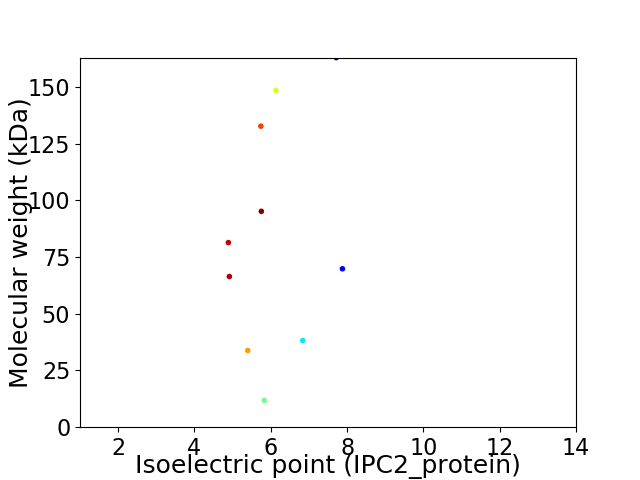
Virtual 2D-PAGE plot for 10 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
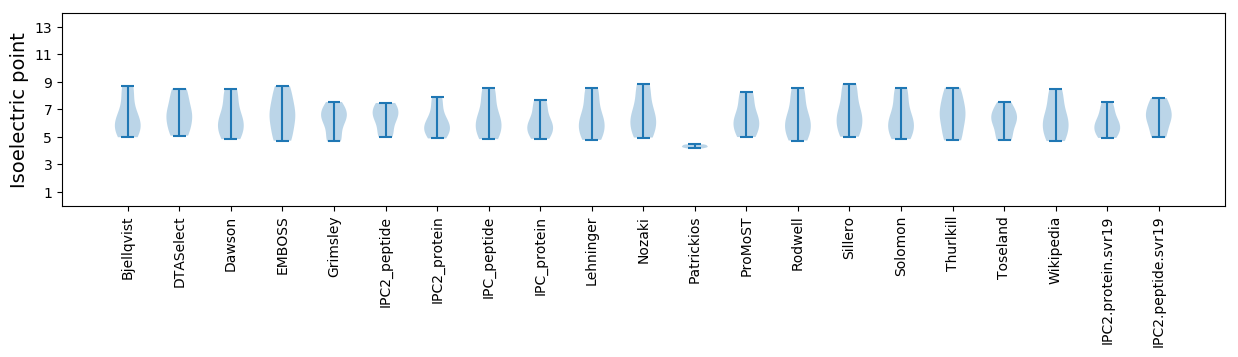
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A291B0C8|A0A291B0C8_9REOV Uncharacterized protein OS=Tai Forest reovirus OX=2039230 PE=4 SV=1
SS1 pKa = 7.29SIRR4 pKa = 11.84LTGDD8 pKa = 4.0TINHH12 pKa = 5.8GLEE15 pKa = 4.68SIAATHH21 pKa = 7.62DD22 pKa = 3.37DD23 pKa = 3.57RR24 pKa = 11.84QFLVEE29 pKa = 4.65LSGKK33 pKa = 9.67HH34 pKa = 4.98VGRR37 pKa = 11.84IQDD40 pKa = 3.29LWSRR44 pKa = 11.84SLPEE48 pKa = 3.44GWLFQVRR55 pKa = 11.84GKK57 pKa = 9.9LFCFIPIDD65 pKa = 3.34IPLFVHH71 pKa = 6.7EE72 pKa = 5.56FYY74 pKa = 10.66QEE76 pKa = 3.63KK77 pKa = 10.31GVIFVRR83 pKa = 11.84VNGLDD88 pKa = 5.44DD89 pKa = 3.79MLSWWAEE96 pKa = 3.5FDD98 pKa = 3.4PAEE101 pKa = 4.68RR102 pKa = 11.84IIHH105 pKa = 5.92CEE107 pKa = 3.65SYY109 pKa = 11.41LNDD112 pKa = 3.09EE113 pKa = 4.08LAYY116 pKa = 10.98NRR118 pKa = 11.84VALTQGRR125 pKa = 11.84AIMFEE130 pKa = 3.88LARR133 pKa = 11.84RR134 pKa = 11.84EE135 pKa = 3.99RR136 pKa = 11.84DD137 pKa = 3.51TVILDD142 pKa = 2.96IHH144 pKa = 6.99ALIIQNAAAAIQGVDD159 pKa = 3.58ADD161 pKa = 4.61DD162 pKa = 4.47MYY164 pKa = 11.66SGLDD168 pKa = 3.57GIIPVRR174 pKa = 11.84DD175 pKa = 3.26EE176 pKa = 3.84GAFRR180 pKa = 11.84GGGPLRR186 pKa = 11.84PAILPGAVAQPPVAGVRR203 pKa = 11.84LKK205 pKa = 10.72ALPPLPVADD214 pKa = 4.05EE215 pKa = 4.19NGLRR219 pKa = 11.84EE220 pKa = 4.36GEE222 pKa = 4.17VDD224 pKa = 4.33LLAHH228 pKa = 5.66QVSNLQVQQGIQQAAAGPQVQDD250 pKa = 3.57VVVDD254 pKa = 3.87PMNVPLPDD262 pKa = 4.83DD263 pKa = 4.37DD264 pKa = 5.51EE265 pKa = 5.98FMDD268 pKa = 3.93VAPAEE273 pKa = 4.35PPQVIPAADD282 pKa = 3.42VAVEE286 pKa = 3.83RR287 pKa = 11.84RR288 pKa = 11.84MPPVVQNVALQVDD301 pKa = 4.63VPPLNDD307 pKa = 3.32RR308 pKa = 11.84ANYY311 pKa = 9.89RR312 pKa = 11.84LRR314 pKa = 11.84NLPYY318 pKa = 10.42QNDD321 pKa = 3.65QIRR324 pKa = 11.84RR325 pKa = 11.84KK326 pKa = 9.12YY327 pKa = 10.38EE328 pKa = 3.62EE329 pKa = 4.23YY330 pKa = 10.82EE331 pKa = 4.52DD332 pKa = 3.51IFEE335 pKa = 4.43TMRR338 pKa = 11.84GAVEE342 pKa = 3.83DD343 pKa = 3.8ANRR346 pKa = 11.84YY347 pKa = 8.56INNIQDD353 pKa = 3.87DD354 pKa = 4.56VLTLEE359 pKa = 4.26NLRR362 pKa = 11.84NIPQAFQILEE372 pKa = 4.1RR373 pKa = 11.84FVVRR377 pKa = 11.84NTARR381 pKa = 11.84LDD383 pKa = 3.52PFRR386 pKa = 11.84LPSSTLNEE394 pKa = 4.23TILEE398 pKa = 4.07QEE400 pKa = 4.19YY401 pKa = 11.1GLLHH405 pKa = 7.66DD406 pKa = 3.97EE407 pKa = 4.57VAWVRR412 pKa = 11.84DD413 pKa = 3.75VGPEE417 pKa = 3.79EE418 pKa = 4.18VDD420 pKa = 3.15PDD422 pKa = 3.9RR423 pKa = 11.84LDD425 pKa = 3.55EE426 pKa = 4.29QIRR429 pKa = 11.84DD430 pKa = 3.57YY431 pKa = 11.38DD432 pKa = 3.55EE433 pKa = 5.39RR434 pKa = 11.84LADD437 pKa = 3.77EE438 pKa = 4.36EE439 pKa = 4.27MALIEE444 pKa = 4.37AEE446 pKa = 4.25SRR448 pKa = 11.84ALEE451 pKa = 4.26AKK453 pKa = 10.19LMRR456 pKa = 11.84VGIPVGAGTGLEE468 pKa = 4.41TIRR471 pKa = 11.84VEE473 pKa = 4.09YY474 pKa = 10.95GMVGAIDD481 pKa = 3.61QGPYY485 pKa = 9.41IDD487 pKa = 5.99DD488 pKa = 3.83YY489 pKa = 12.21VEE491 pKa = 5.55DD492 pKa = 5.71DD493 pKa = 3.74VLSWTGFYY501 pKa = 10.91QNFSLDD507 pKa = 3.6KK508 pKa = 10.02VFHH511 pKa = 7.11DD512 pKa = 4.67FFSRR516 pKa = 11.84IPDD519 pKa = 3.24IPTGRR524 pKa = 11.84RR525 pKa = 11.84MLARR529 pKa = 11.84VMRR532 pKa = 11.84DD533 pKa = 2.83VQTRR537 pKa = 11.84VSTLTSVNRR546 pKa = 11.84EE547 pKa = 3.71RR548 pKa = 11.84FRR550 pKa = 11.84EE551 pKa = 3.96PNGIRR556 pKa = 11.84DD557 pKa = 3.65LAGVSIVPGTAFNMMLRR574 pKa = 11.84WSTLSARR581 pKa = 11.84FAAGGRR587 pKa = 11.84HH588 pKa = 4.71RR589 pKa = 11.84QVCMEE594 pKa = 4.53LARR597 pKa = 11.84RR598 pKa = 11.84YY599 pKa = 10.09AGWRR603 pKa = 11.84AGARR607 pKa = 11.84YY608 pKa = 7.32TIPDD612 pKa = 3.49GLLRR616 pKa = 11.84ALRR619 pKa = 11.84NHH621 pKa = 6.97RR622 pKa = 11.84LHH624 pKa = 6.65TYY626 pKa = 9.58VPLGRR631 pKa = 11.84IYY633 pKa = 10.41IAVHH637 pKa = 6.36HH638 pKa = 5.77MMEE641 pKa = 4.11GRR643 pKa = 11.84EE644 pKa = 3.94NQAGDD649 pKa = 3.12IRR651 pKa = 11.84SIVMDD656 pKa = 4.89RR657 pKa = 11.84NRR659 pKa = 11.84ADD661 pKa = 2.79QWRR664 pKa = 11.84RR665 pKa = 11.84IHH667 pKa = 6.94PSLPYY672 pKa = 10.55VMHH675 pKa = 6.8IEE677 pKa = 4.04FWEE680 pKa = 4.19MFVDD684 pKa = 3.83WFPDD688 pKa = 3.11VGTGGIVNMSPIFPTSLSDD707 pKa = 3.59RR708 pKa = 11.84EE709 pKa = 4.09VWVAARR715 pKa = 11.84LLL717 pKa = 3.9
SS1 pKa = 7.29SIRR4 pKa = 11.84LTGDD8 pKa = 4.0TINHH12 pKa = 5.8GLEE15 pKa = 4.68SIAATHH21 pKa = 7.62DD22 pKa = 3.37DD23 pKa = 3.57RR24 pKa = 11.84QFLVEE29 pKa = 4.65LSGKK33 pKa = 9.67HH34 pKa = 4.98VGRR37 pKa = 11.84IQDD40 pKa = 3.29LWSRR44 pKa = 11.84SLPEE48 pKa = 3.44GWLFQVRR55 pKa = 11.84GKK57 pKa = 9.9LFCFIPIDD65 pKa = 3.34IPLFVHH71 pKa = 6.7EE72 pKa = 5.56FYY74 pKa = 10.66QEE76 pKa = 3.63KK77 pKa = 10.31GVIFVRR83 pKa = 11.84VNGLDD88 pKa = 5.44DD89 pKa = 3.79MLSWWAEE96 pKa = 3.5FDD98 pKa = 3.4PAEE101 pKa = 4.68RR102 pKa = 11.84IIHH105 pKa = 5.92CEE107 pKa = 3.65SYY109 pKa = 11.41LNDD112 pKa = 3.09EE113 pKa = 4.08LAYY116 pKa = 10.98NRR118 pKa = 11.84VALTQGRR125 pKa = 11.84AIMFEE130 pKa = 3.88LARR133 pKa = 11.84RR134 pKa = 11.84EE135 pKa = 3.99RR136 pKa = 11.84DD137 pKa = 3.51TVILDD142 pKa = 2.96IHH144 pKa = 6.99ALIIQNAAAAIQGVDD159 pKa = 3.58ADD161 pKa = 4.61DD162 pKa = 4.47MYY164 pKa = 11.66SGLDD168 pKa = 3.57GIIPVRR174 pKa = 11.84DD175 pKa = 3.26EE176 pKa = 3.84GAFRR180 pKa = 11.84GGGPLRR186 pKa = 11.84PAILPGAVAQPPVAGVRR203 pKa = 11.84LKK205 pKa = 10.72ALPPLPVADD214 pKa = 4.05EE215 pKa = 4.19NGLRR219 pKa = 11.84EE220 pKa = 4.36GEE222 pKa = 4.17VDD224 pKa = 4.33LLAHH228 pKa = 5.66QVSNLQVQQGIQQAAAGPQVQDD250 pKa = 3.57VVVDD254 pKa = 3.87PMNVPLPDD262 pKa = 4.83DD263 pKa = 4.37DD264 pKa = 5.51EE265 pKa = 5.98FMDD268 pKa = 3.93VAPAEE273 pKa = 4.35PPQVIPAADD282 pKa = 3.42VAVEE286 pKa = 3.83RR287 pKa = 11.84RR288 pKa = 11.84MPPVVQNVALQVDD301 pKa = 4.63VPPLNDD307 pKa = 3.32RR308 pKa = 11.84ANYY311 pKa = 9.89RR312 pKa = 11.84LRR314 pKa = 11.84NLPYY318 pKa = 10.42QNDD321 pKa = 3.65QIRR324 pKa = 11.84RR325 pKa = 11.84KK326 pKa = 9.12YY327 pKa = 10.38EE328 pKa = 3.62EE329 pKa = 4.23YY330 pKa = 10.82EE331 pKa = 4.52DD332 pKa = 3.51IFEE335 pKa = 4.43TMRR338 pKa = 11.84GAVEE342 pKa = 3.83DD343 pKa = 3.8ANRR346 pKa = 11.84YY347 pKa = 8.56INNIQDD353 pKa = 3.87DD354 pKa = 4.56VLTLEE359 pKa = 4.26NLRR362 pKa = 11.84NIPQAFQILEE372 pKa = 4.1RR373 pKa = 11.84FVVRR377 pKa = 11.84NTARR381 pKa = 11.84LDD383 pKa = 3.52PFRR386 pKa = 11.84LPSSTLNEE394 pKa = 4.23TILEE398 pKa = 4.07QEE400 pKa = 4.19YY401 pKa = 11.1GLLHH405 pKa = 7.66DD406 pKa = 3.97EE407 pKa = 4.57VAWVRR412 pKa = 11.84DD413 pKa = 3.75VGPEE417 pKa = 3.79EE418 pKa = 4.18VDD420 pKa = 3.15PDD422 pKa = 3.9RR423 pKa = 11.84LDD425 pKa = 3.55EE426 pKa = 4.29QIRR429 pKa = 11.84DD430 pKa = 3.57YY431 pKa = 11.38DD432 pKa = 3.55EE433 pKa = 5.39RR434 pKa = 11.84LADD437 pKa = 3.77EE438 pKa = 4.36EE439 pKa = 4.27MALIEE444 pKa = 4.37AEE446 pKa = 4.25SRR448 pKa = 11.84ALEE451 pKa = 4.26AKK453 pKa = 10.19LMRR456 pKa = 11.84VGIPVGAGTGLEE468 pKa = 4.41TIRR471 pKa = 11.84VEE473 pKa = 4.09YY474 pKa = 10.95GMVGAIDD481 pKa = 3.61QGPYY485 pKa = 9.41IDD487 pKa = 5.99DD488 pKa = 3.83YY489 pKa = 12.21VEE491 pKa = 5.55DD492 pKa = 5.71DD493 pKa = 3.74VLSWTGFYY501 pKa = 10.91QNFSLDD507 pKa = 3.6KK508 pKa = 10.02VFHH511 pKa = 7.11DD512 pKa = 4.67FFSRR516 pKa = 11.84IPDD519 pKa = 3.24IPTGRR524 pKa = 11.84RR525 pKa = 11.84MLARR529 pKa = 11.84VMRR532 pKa = 11.84DD533 pKa = 2.83VQTRR537 pKa = 11.84VSTLTSVNRR546 pKa = 11.84EE547 pKa = 3.71RR548 pKa = 11.84FRR550 pKa = 11.84EE551 pKa = 3.96PNGIRR556 pKa = 11.84DD557 pKa = 3.65LAGVSIVPGTAFNMMLRR574 pKa = 11.84WSTLSARR581 pKa = 11.84FAAGGRR587 pKa = 11.84HH588 pKa = 4.71RR589 pKa = 11.84QVCMEE594 pKa = 4.53LARR597 pKa = 11.84RR598 pKa = 11.84YY599 pKa = 10.09AGWRR603 pKa = 11.84AGARR607 pKa = 11.84YY608 pKa = 7.32TIPDD612 pKa = 3.49GLLRR616 pKa = 11.84ALRR619 pKa = 11.84NHH621 pKa = 6.97RR622 pKa = 11.84LHH624 pKa = 6.65TYY626 pKa = 9.58VPLGRR631 pKa = 11.84IYY633 pKa = 10.41IAVHH637 pKa = 6.36HH638 pKa = 5.77MMEE641 pKa = 4.11GRR643 pKa = 11.84EE644 pKa = 3.94NQAGDD649 pKa = 3.12IRR651 pKa = 11.84SIVMDD656 pKa = 4.89RR657 pKa = 11.84NRR659 pKa = 11.84ADD661 pKa = 2.79QWRR664 pKa = 11.84RR665 pKa = 11.84IHH667 pKa = 6.94PSLPYY672 pKa = 10.55VMHH675 pKa = 6.8IEE677 pKa = 4.04FWEE680 pKa = 4.19MFVDD684 pKa = 3.83WFPDD688 pKa = 3.11VGTGGIVNMSPIFPTSLSDD707 pKa = 3.59RR708 pKa = 11.84EE709 pKa = 4.09VWVAARR715 pKa = 11.84LLL717 pKa = 3.9
Molecular weight: 81.35 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A291ID07|A0A291ID07_9REOV Putative membrane-spanning protein (Fragment) OS=Tai Forest reovirus OX=2039230 PE=4 SV=1
LL1 pKa = 7.22RR2 pKa = 11.84RR3 pKa = 11.84PVKK6 pKa = 10.79YY7 pKa = 10.67NIFLLPTARR16 pKa = 11.84KK17 pKa = 8.64LAQKK21 pKa = 10.52LKK23 pKa = 11.03LFGKK27 pKa = 10.2LDD29 pKa = 3.89RR30 pKa = 11.84LPPEE34 pKa = 4.84DD35 pKa = 3.33WRR37 pKa = 11.84SWGALEE43 pKa = 6.28SKK45 pKa = 10.33LKK47 pKa = 10.14WLINTYY53 pKa = 9.68PSNWRR58 pKa = 11.84EE59 pKa = 3.85KK60 pKa = 10.22ISAHH64 pKa = 5.37EE65 pKa = 4.05RR66 pKa = 11.84GGMKK70 pKa = 10.23FAIQVEE76 pKa = 4.48KK77 pKa = 11.11GSDD80 pKa = 3.19VDD82 pKa = 4.11SAKK85 pKa = 9.9STIRR89 pKa = 11.84KK90 pKa = 9.28LMAQYY95 pKa = 10.79GKK97 pKa = 10.53RR98 pKa = 11.84INDD101 pKa = 3.53LQFVGVDD108 pKa = 3.61DD109 pKa = 4.86LKK111 pKa = 11.44NRR113 pKa = 11.84YY114 pKa = 8.92KK115 pKa = 10.49KK116 pKa = 10.8GKK118 pKa = 8.98IVLSVSEE125 pKa = 5.22RR126 pKa = 11.84IDD128 pKa = 3.4EE129 pKa = 4.27SAEE132 pKa = 4.29GILMKK137 pKa = 10.02WRR139 pKa = 11.84EE140 pKa = 4.01ANQDD144 pKa = 3.22QFGKK148 pKa = 11.17YY149 pKa = 7.88MGEE152 pKa = 3.78MLKK155 pKa = 10.7PKK157 pKa = 9.91MLVTDD162 pKa = 5.18FINPQAGCRR171 pKa = 11.84YY172 pKa = 8.76PNVDD176 pKa = 4.21ALSEE180 pKa = 4.58LIPEE184 pKa = 4.42DD185 pKa = 4.55SSWLFQAQAGWQMTYY200 pKa = 10.33GHH202 pKa = 7.39LRR204 pKa = 11.84PLEE207 pKa = 4.76IGMKK211 pKa = 9.84EE212 pKa = 4.02DD213 pKa = 5.19FFASSCDD220 pKa = 3.07IWCVRR225 pKa = 11.84PFSIYY230 pKa = 9.92PKK232 pKa = 10.87YY233 pKa = 10.5EE234 pKa = 3.81GLPQNFSGALAVLPTNLLTDD254 pKa = 4.27PEE256 pKa = 4.42VKK258 pKa = 10.3LQSEE262 pKa = 4.75LMSKK266 pKa = 10.82AKK268 pKa = 10.45GSSKK272 pKa = 11.16DD273 pKa = 3.48NIVFCFSYY281 pKa = 10.97LSFTFRR287 pKa = 11.84SDD289 pKa = 3.18EE290 pKa = 4.24ALSTMKK296 pKa = 10.88KK297 pKa = 10.24LGLPLAEE304 pKa = 4.14KK305 pKa = 10.09EE306 pKa = 4.53VYY308 pKa = 8.92TVLVPKK314 pKa = 10.56SVIPPLGPISGEE326 pKa = 3.81RR327 pKa = 11.84FAKK330 pKa = 10.38DD331 pKa = 3.23VLSRR335 pKa = 11.84AVGAVNTKK343 pKa = 10.49SKK345 pKa = 10.73IAMTFASGLNPDD357 pKa = 3.62VTVLCEE363 pKa = 4.16TGCGTFIDD371 pKa = 4.46GTNPRR376 pKa = 11.84MMAEE380 pKa = 4.11ILSEE384 pKa = 4.07PARR387 pKa = 11.84MIILGRR393 pKa = 11.84KK394 pKa = 8.94GGGKK398 pKa = 9.45SRR400 pKa = 11.84LGALFSEE407 pKa = 4.9LGYY410 pKa = 11.09NVIDD414 pKa = 3.55SDD416 pKa = 4.18SYY418 pKa = 11.57GRR420 pKa = 11.84LLYY423 pKa = 10.09MCGADD428 pKa = 3.35MNEE431 pKa = 3.98EE432 pKa = 4.25KK433 pKa = 10.3FLEE436 pKa = 4.71MIDD439 pKa = 3.35VYY441 pKa = 11.59LRR443 pKa = 11.84MTKK446 pKa = 9.93KK447 pKa = 10.39EE448 pKa = 3.93RR449 pKa = 11.84DD450 pKa = 3.63SVPSYY455 pKa = 11.23FEE457 pKa = 4.19VEE459 pKa = 3.74MDD461 pKa = 5.31KK462 pKa = 10.53MLQHH466 pKa = 4.75YY467 pKa = 7.03TEE469 pKa = 4.39WLGLPHH475 pKa = 6.29YY476 pKa = 10.32LSRR479 pKa = 11.84EE480 pKa = 3.67HH481 pKa = 7.44DD482 pKa = 3.48SYY484 pKa = 12.07GRR486 pKa = 11.84LDD488 pKa = 3.22RR489 pKa = 11.84ALFHH493 pKa = 6.99SFDD496 pKa = 4.32KK497 pKa = 10.8IYY499 pKa = 10.98DD500 pKa = 3.49SALRR504 pKa = 11.84KK505 pKa = 9.85VSPEE509 pKa = 3.55FMRR512 pKa = 11.84LNYY515 pKa = 8.68TKK517 pKa = 10.56RR518 pKa = 11.84LWEE521 pKa = 4.11GVGFDD526 pKa = 4.93DD527 pKa = 5.76NGIPSTGRR535 pKa = 11.84EE536 pKa = 3.52RR537 pKa = 11.84YY538 pKa = 9.51VCFCHH543 pKa = 6.44TLIEE547 pKa = 4.35QFQGMSATMFEE558 pKa = 5.6LIPSHH563 pKa = 5.25STRR566 pKa = 11.84VAISLRR572 pKa = 11.84GQGTSLNSEE581 pKa = 4.95LYY583 pKa = 10.0LHH585 pKa = 7.44DD586 pKa = 4.26YY587 pKa = 10.56YY588 pKa = 11.41VAKK591 pKa = 10.64NGNGCTKK598 pKa = 10.53VGLGWLVYY606 pKa = 9.95MLEE609 pKa = 4.16QKK611 pKa = 9.77MKK613 pKa = 10.18EE614 pKa = 3.87
LL1 pKa = 7.22RR2 pKa = 11.84RR3 pKa = 11.84PVKK6 pKa = 10.79YY7 pKa = 10.67NIFLLPTARR16 pKa = 11.84KK17 pKa = 8.64LAQKK21 pKa = 10.52LKK23 pKa = 11.03LFGKK27 pKa = 10.2LDD29 pKa = 3.89RR30 pKa = 11.84LPPEE34 pKa = 4.84DD35 pKa = 3.33WRR37 pKa = 11.84SWGALEE43 pKa = 6.28SKK45 pKa = 10.33LKK47 pKa = 10.14WLINTYY53 pKa = 9.68PSNWRR58 pKa = 11.84EE59 pKa = 3.85KK60 pKa = 10.22ISAHH64 pKa = 5.37EE65 pKa = 4.05RR66 pKa = 11.84GGMKK70 pKa = 10.23FAIQVEE76 pKa = 4.48KK77 pKa = 11.11GSDD80 pKa = 3.19VDD82 pKa = 4.11SAKK85 pKa = 9.9STIRR89 pKa = 11.84KK90 pKa = 9.28LMAQYY95 pKa = 10.79GKK97 pKa = 10.53RR98 pKa = 11.84INDD101 pKa = 3.53LQFVGVDD108 pKa = 3.61DD109 pKa = 4.86LKK111 pKa = 11.44NRR113 pKa = 11.84YY114 pKa = 8.92KK115 pKa = 10.49KK116 pKa = 10.8GKK118 pKa = 8.98IVLSVSEE125 pKa = 5.22RR126 pKa = 11.84IDD128 pKa = 3.4EE129 pKa = 4.27SAEE132 pKa = 4.29GILMKK137 pKa = 10.02WRR139 pKa = 11.84EE140 pKa = 4.01ANQDD144 pKa = 3.22QFGKK148 pKa = 11.17YY149 pKa = 7.88MGEE152 pKa = 3.78MLKK155 pKa = 10.7PKK157 pKa = 9.91MLVTDD162 pKa = 5.18FINPQAGCRR171 pKa = 11.84YY172 pKa = 8.76PNVDD176 pKa = 4.21ALSEE180 pKa = 4.58LIPEE184 pKa = 4.42DD185 pKa = 4.55SSWLFQAQAGWQMTYY200 pKa = 10.33GHH202 pKa = 7.39LRR204 pKa = 11.84PLEE207 pKa = 4.76IGMKK211 pKa = 9.84EE212 pKa = 4.02DD213 pKa = 5.19FFASSCDD220 pKa = 3.07IWCVRR225 pKa = 11.84PFSIYY230 pKa = 9.92PKK232 pKa = 10.87YY233 pKa = 10.5EE234 pKa = 3.81GLPQNFSGALAVLPTNLLTDD254 pKa = 4.27PEE256 pKa = 4.42VKK258 pKa = 10.3LQSEE262 pKa = 4.75LMSKK266 pKa = 10.82AKK268 pKa = 10.45GSSKK272 pKa = 11.16DD273 pKa = 3.48NIVFCFSYY281 pKa = 10.97LSFTFRR287 pKa = 11.84SDD289 pKa = 3.18EE290 pKa = 4.24ALSTMKK296 pKa = 10.88KK297 pKa = 10.24LGLPLAEE304 pKa = 4.14KK305 pKa = 10.09EE306 pKa = 4.53VYY308 pKa = 8.92TVLVPKK314 pKa = 10.56SVIPPLGPISGEE326 pKa = 3.81RR327 pKa = 11.84FAKK330 pKa = 10.38DD331 pKa = 3.23VLSRR335 pKa = 11.84AVGAVNTKK343 pKa = 10.49SKK345 pKa = 10.73IAMTFASGLNPDD357 pKa = 3.62VTVLCEE363 pKa = 4.16TGCGTFIDD371 pKa = 4.46GTNPRR376 pKa = 11.84MMAEE380 pKa = 4.11ILSEE384 pKa = 4.07PARR387 pKa = 11.84MIILGRR393 pKa = 11.84KK394 pKa = 8.94GGGKK398 pKa = 9.45SRR400 pKa = 11.84LGALFSEE407 pKa = 4.9LGYY410 pKa = 11.09NVIDD414 pKa = 3.55SDD416 pKa = 4.18SYY418 pKa = 11.57GRR420 pKa = 11.84LLYY423 pKa = 10.09MCGADD428 pKa = 3.35MNEE431 pKa = 3.98EE432 pKa = 4.25KK433 pKa = 10.3FLEE436 pKa = 4.71MIDD439 pKa = 3.35VYY441 pKa = 11.59LRR443 pKa = 11.84MTKK446 pKa = 9.93KK447 pKa = 10.39EE448 pKa = 3.93RR449 pKa = 11.84DD450 pKa = 3.63SVPSYY455 pKa = 11.23FEE457 pKa = 4.19VEE459 pKa = 3.74MDD461 pKa = 5.31KK462 pKa = 10.53MLQHH466 pKa = 4.75YY467 pKa = 7.03TEE469 pKa = 4.39WLGLPHH475 pKa = 6.29YY476 pKa = 10.32LSRR479 pKa = 11.84EE480 pKa = 3.67HH481 pKa = 7.44DD482 pKa = 3.48SYY484 pKa = 12.07GRR486 pKa = 11.84LDD488 pKa = 3.22RR489 pKa = 11.84ALFHH493 pKa = 6.99SFDD496 pKa = 4.32KK497 pKa = 10.8IYY499 pKa = 10.98DD500 pKa = 3.49SALRR504 pKa = 11.84KK505 pKa = 9.85VSPEE509 pKa = 3.55FMRR512 pKa = 11.84LNYY515 pKa = 8.68TKK517 pKa = 10.56RR518 pKa = 11.84LWEE521 pKa = 4.11GVGFDD526 pKa = 4.93DD527 pKa = 5.76NGIPSTGRR535 pKa = 11.84EE536 pKa = 3.52RR537 pKa = 11.84YY538 pKa = 9.51VCFCHH543 pKa = 6.44TLIEE547 pKa = 4.35QFQGMSATMFEE558 pKa = 5.6LIPSHH563 pKa = 5.25STRR566 pKa = 11.84VAISLRR572 pKa = 11.84GQGTSLNSEE581 pKa = 4.95LYY583 pKa = 10.0LHH585 pKa = 7.44DD586 pKa = 4.26YY587 pKa = 10.56YY588 pKa = 11.41VAKK591 pKa = 10.64NGNGCTKK598 pKa = 10.53VGLGWLVYY606 pKa = 9.95MLEE609 pKa = 4.16QKK611 pKa = 9.77MKK613 pKa = 10.18EE614 pKa = 3.87
Molecular weight: 69.81 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
7421 |
107 |
1433 |
742.1 |
84.05 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.926 ± 0.36 | 1.415 ± 0.185 |
6.279 ± 0.353 | 6.926 ± 0.361 |
4.299 ± 0.164 | 7.533 ± 0.4 |
2.21 ± 0.2 | 5.363 ± 0.333 |
4.609 ± 0.619 | 9.069 ± 0.228 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.611 ± 0.105 | 3.584 ± 0.292 |
4.77 ± 0.302 | 3.059 ± 0.164 |
7.492 ± 0.441 | 5.511 ± 0.352 |
4.42 ± 0.381 | 7.775 ± 0.249 |
1.711 ± 0.121 | 3.396 ± 0.24 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
