
Bat coronavirus HKU5 (BtCoV) (BtCoV/HKU5/2004)
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Pisoniviricetes; Nidovirales; Cornidovirineae; Coronaviridae; Orthocoronavirinae; Betacoronavirus; Merbecovirus
Average proteome isoelectric point is 7.25
Get precalculated fractions of proteins
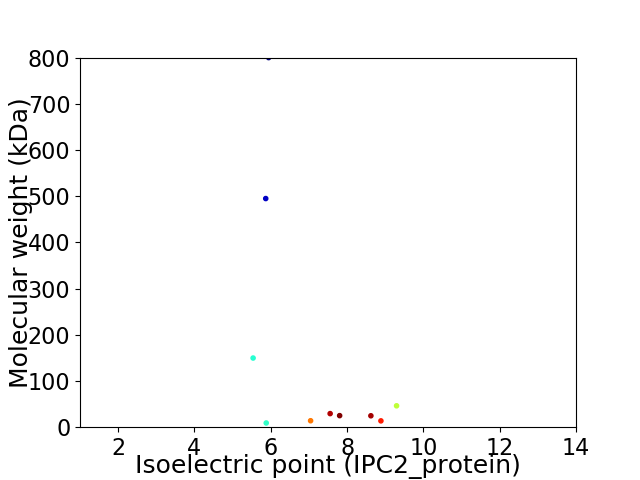
Virtual 2D-PAGE plot for 10 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|A3EXD1|NS3A_BCHK5 Non-structural protein 3a OS=Bat coronavirus HKU5 OX=694008 GN=3a PE=3 SV=1
MM1 pKa = 7.16IRR3 pKa = 11.84SVLVLMCSLTFIGNLTRR20 pKa = 11.84GQSVDD25 pKa = 3.09MGHH28 pKa = 6.56NGTGSCLDD36 pKa = 3.67SQVQPDD42 pKa = 4.15YY43 pKa = 11.05FEE45 pKa = 5.36SVHH48 pKa = 5.43TTWPMPIDD56 pKa = 3.44TSKK59 pKa = 11.52AEE61 pKa = 4.0GVIYY65 pKa = 10.04PNGKK69 pKa = 9.38SYY71 pKa = 11.79SNITLTYY78 pKa = 8.48TGLYY82 pKa = 9.29PKK84 pKa = 10.87ANDD87 pKa = 3.99LGKK90 pKa = 10.44QYY92 pKa = 10.84LFSDD96 pKa = 4.14GHH98 pKa = 5.51SAPGRR103 pKa = 11.84LNNLFVSNYY112 pKa = 8.4SSQVEE117 pKa = 4.3SFDD120 pKa = 3.77DD121 pKa = 4.42GFVVRR126 pKa = 11.84IGAAANKK133 pKa = 9.28TGTTVISQSTFKK145 pKa = 10.09PIKK148 pKa = 9.78KK149 pKa = 9.62IYY151 pKa = 8.89PAFLLGHH158 pKa = 5.68SVGNYY163 pKa = 7.01TPSNRR168 pKa = 11.84TGRR171 pKa = 11.84YY172 pKa = 8.95LNHH175 pKa = 6.74TLVILPDD182 pKa = 3.55GCGTILHH189 pKa = 7.04AFYY192 pKa = 10.81CVLHH196 pKa = 6.74PRR198 pKa = 11.84TQQNCAGEE206 pKa = 4.56TNFKK210 pKa = 10.74SLSLWDD216 pKa = 4.4TPASDD221 pKa = 4.53CVSGSYY227 pKa = 9.41NQEE230 pKa = 3.54ATLGAFKK237 pKa = 10.95VYY239 pKa = 10.44FDD241 pKa = 6.08LINCTFRR248 pKa = 11.84YY249 pKa = 10.19NYY251 pKa = 9.77TITEE255 pKa = 4.22DD256 pKa = 4.48EE257 pKa = 3.97NAEE260 pKa = 4.14WFGITQDD267 pKa = 3.47TQGVHH272 pKa = 6.7LYY274 pKa = 10.45SSRR277 pKa = 11.84KK278 pKa = 9.46EE279 pKa = 3.81NVFRR283 pKa = 11.84NNMFHH288 pKa = 7.46FATLPVYY295 pKa = 10.33QKK297 pKa = 10.09ILYY300 pKa = 7.25YY301 pKa = 9.63TVIPRR306 pKa = 11.84SIRR309 pKa = 11.84SPFNDD314 pKa = 2.95RR315 pKa = 11.84KK316 pKa = 10.37AWAAFYY322 pKa = 9.99IYY324 pKa = 10.33KK325 pKa = 10.23LHH327 pKa = 7.02PLTYY331 pKa = 10.54LLNFDD336 pKa = 3.62VEE338 pKa = 4.81GYY340 pKa = 7.31ITKK343 pKa = 10.51AVDD346 pKa = 3.43CGYY349 pKa = 11.23DD350 pKa = 4.06DD351 pKa = 5.81LAQLQCSYY359 pKa = 11.37EE360 pKa = 4.24SFEE363 pKa = 4.45VEE365 pKa = 3.78TGVYY369 pKa = 9.84SVSSFEE375 pKa = 3.78ASPRR379 pKa = 11.84GEE381 pKa = 4.58FIEE384 pKa = 4.21QATTQEE390 pKa = 4.29CDD392 pKa = 3.74FTPMLTGTPPPIYY405 pKa = 9.95NFKK408 pKa = 10.75RR409 pKa = 11.84LVFTNCNYY417 pKa = 10.82NLTKK421 pKa = 10.58LLSLFQVSEE430 pKa = 4.71FSCHH434 pKa = 4.9QVSPSSLATGCYY446 pKa = 9.95SSLTVDD452 pKa = 3.43YY453 pKa = 9.87FAYY456 pKa = 8.82STDD459 pKa = 3.07MSSYY463 pKa = 9.68LQPGSAGAIVQFNYY477 pKa = 10.33KK478 pKa = 9.78QDD480 pKa = 3.66FSNPTCRR487 pKa = 11.84VLATVPQNLTTITKK501 pKa = 8.73PSNYY505 pKa = 10.1AYY507 pKa = 9.51LTEE510 pKa = 4.67CYY512 pKa = 7.89KK513 pKa = 10.6TSAYY517 pKa = 9.25GKK519 pKa = 9.32NYY521 pKa = 10.18LYY523 pKa = 10.26NAPGAYY529 pKa = 8.3TPCLSLASRR538 pKa = 11.84GFSTKK543 pKa = 10.14YY544 pKa = 9.57QSHH547 pKa = 6.82SDD549 pKa = 3.76GEE551 pKa = 4.52LTTTGYY557 pKa = 10.29IYY559 pKa = 10.33PVTGNLQMAFIISVQYY575 pKa = 9.07GTDD578 pKa = 3.55TNSVCPMQALRR589 pKa = 11.84NDD591 pKa = 3.48TSIEE595 pKa = 4.13DD596 pKa = 3.73KK597 pKa = 11.26LDD599 pKa = 3.35VCVEE603 pKa = 3.97YY604 pKa = 10.66SLHH607 pKa = 6.69GITGRR612 pKa = 11.84GVFHH616 pKa = 7.07NCTSVGLRR624 pKa = 11.84NQRR627 pKa = 11.84FVYY630 pKa = 9.22DD631 pKa = 3.57TFDD634 pKa = 3.52NLVGYY639 pKa = 10.57HH640 pKa = 6.66SDD642 pKa = 3.15NGNYY646 pKa = 9.55YY647 pKa = 9.96CVRR650 pKa = 11.84PCVSVPVSVIYY661 pKa = 10.76DD662 pKa = 3.52KK663 pKa = 11.4ASNSHH668 pKa = 5.69ATLFGSVACSHH679 pKa = 5.84VTTMMSQFSRR689 pKa = 11.84MTKK692 pKa = 9.37TNLLARR698 pKa = 11.84TTPGPLQTTVGCAMGFINSSMVVDD722 pKa = 4.66EE723 pKa = 4.72CQLPLGQSLCAIPPTTSSRR742 pKa = 11.84VRR744 pKa = 11.84RR745 pKa = 11.84ATSGASDD752 pKa = 3.62VFQIATLNFTSPLTLAPINSTGFVVAVPTNFTFGVTQEE790 pKa = 4.56FIEE793 pKa = 4.32TTIQKK798 pKa = 8.34ITVDD802 pKa = 3.89CKK804 pKa = 10.81QYY806 pKa = 10.53VCNGFKK812 pKa = 10.45KK813 pKa = 10.92CEE815 pKa = 4.15DD816 pKa = 3.47LLKK819 pKa = 10.63EE820 pKa = 4.03YY821 pKa = 10.97GQFCSKK827 pKa = 10.24INQALHH833 pKa = 5.92GANLRR838 pKa = 11.84QDD840 pKa = 3.5EE841 pKa = 4.74SIANLFSSIKK851 pKa = 8.36TQNTQPLQAGLNGDD865 pKa = 4.19FNLTMLQIPQVTTGEE880 pKa = 3.87RR881 pKa = 11.84KK882 pKa = 9.32YY883 pKa = 10.94RR884 pKa = 11.84STIEE888 pKa = 4.38DD889 pKa = 3.78LLFNKK894 pKa = 7.82VTIADD899 pKa = 3.7PGYY902 pKa = 7.86MQGYY906 pKa = 8.62DD907 pKa = 2.97EE908 pKa = 5.92CMQQGPQSARR918 pKa = 11.84DD919 pKa = 4.21LICAQYY925 pKa = 9.2VAGYY929 pKa = 9.86KK930 pKa = 9.71VLPPLYY936 pKa = 9.9DD937 pKa = 3.96PYY939 pKa = 10.78MEE941 pKa = 4.81AAYY944 pKa = 9.18TSSLLGSIAGASWTAGLSSFAAIPFAQSIFYY975 pKa = 10.26RR976 pKa = 11.84LNGVGITQQVLSEE989 pKa = 4.05NQKK992 pKa = 10.46IIANKK997 pKa = 9.42FNQALGAMQTGFTTTNLAFNKK1018 pKa = 9.95VQDD1021 pKa = 3.97AVNANAMALSKK1032 pKa = 10.7LAAEE1036 pKa = 4.98LSNTFGAISSSISDD1050 pKa = 3.1ILARR1054 pKa = 11.84LDD1056 pKa = 3.62TVEE1059 pKa = 5.08QEE1061 pKa = 4.13AQIDD1065 pKa = 3.71RR1066 pKa = 11.84LINGRR1071 pKa = 11.84LTSLNAFVAQQLVRR1085 pKa = 11.84TEE1087 pKa = 3.65AAARR1091 pKa = 11.84SAQLAQDD1098 pKa = 3.82KK1099 pKa = 11.07VNEE1102 pKa = 4.39CVKK1105 pKa = 10.74SQSKK1109 pKa = 10.87RR1110 pKa = 11.84NGFCGTGTHH1119 pKa = 5.85IVSFAINAPNGLYY1132 pKa = 10.03FFHH1135 pKa = 7.75VGYY1138 pKa = 10.58QPTSHH1143 pKa = 6.33VNATAAYY1150 pKa = 8.36GLCNTEE1156 pKa = 4.45NPQKK1160 pKa = 10.6CIAPIDD1166 pKa = 4.22GYY1168 pKa = 11.21FVLNQTTSTVADD1180 pKa = 4.45SDD1182 pKa = 3.99QQWYY1186 pKa = 7.3YY1187 pKa = 9.08TGSSFFHH1194 pKa = 6.96PEE1196 pKa = 4.31PITEE1200 pKa = 4.24ANSKK1204 pKa = 8.77YY1205 pKa = 11.08VSMDD1209 pKa = 3.11VKK1211 pKa = 11.03FEE1213 pKa = 4.13NLTNRR1218 pKa = 11.84LPPPLLSNSTDD1229 pKa = 3.39LDD1231 pKa = 3.81FKK1233 pKa = 11.54EE1234 pKa = 4.12EE1235 pKa = 4.03LEE1237 pKa = 4.22EE1238 pKa = 4.02FFKK1241 pKa = 11.25NVSSQGPNFQEE1252 pKa = 3.42ISKK1255 pKa = 10.78INTTLLNLNTEE1266 pKa = 4.26LMVLSEE1272 pKa = 4.38VVKK1275 pKa = 10.44QLNEE1279 pKa = 3.95SYY1281 pKa = 10.82IDD1283 pKa = 3.87LKK1285 pKa = 11.29EE1286 pKa = 4.03LGNYY1290 pKa = 7.36TFYY1293 pKa = 11.09QKK1295 pKa = 10.17WPWYY1299 pKa = 8.52IWLGFIAGLVALALCVFFILCCTGCGTSCLGKK1331 pKa = 10.25LKK1333 pKa = 10.69CNRR1336 pKa = 11.84CCDD1339 pKa = 3.46SYY1341 pKa = 12.06DD1342 pKa = 3.43EE1343 pKa = 4.74YY1344 pKa = 10.97EE1345 pKa = 4.0VEE1347 pKa = 4.86KK1348 pKa = 10.84IHH1350 pKa = 6.28VHH1352 pKa = 5.56
MM1 pKa = 7.16IRR3 pKa = 11.84SVLVLMCSLTFIGNLTRR20 pKa = 11.84GQSVDD25 pKa = 3.09MGHH28 pKa = 6.56NGTGSCLDD36 pKa = 3.67SQVQPDD42 pKa = 4.15YY43 pKa = 11.05FEE45 pKa = 5.36SVHH48 pKa = 5.43TTWPMPIDD56 pKa = 3.44TSKK59 pKa = 11.52AEE61 pKa = 4.0GVIYY65 pKa = 10.04PNGKK69 pKa = 9.38SYY71 pKa = 11.79SNITLTYY78 pKa = 8.48TGLYY82 pKa = 9.29PKK84 pKa = 10.87ANDD87 pKa = 3.99LGKK90 pKa = 10.44QYY92 pKa = 10.84LFSDD96 pKa = 4.14GHH98 pKa = 5.51SAPGRR103 pKa = 11.84LNNLFVSNYY112 pKa = 8.4SSQVEE117 pKa = 4.3SFDD120 pKa = 3.77DD121 pKa = 4.42GFVVRR126 pKa = 11.84IGAAANKK133 pKa = 9.28TGTTVISQSTFKK145 pKa = 10.09PIKK148 pKa = 9.78KK149 pKa = 9.62IYY151 pKa = 8.89PAFLLGHH158 pKa = 5.68SVGNYY163 pKa = 7.01TPSNRR168 pKa = 11.84TGRR171 pKa = 11.84YY172 pKa = 8.95LNHH175 pKa = 6.74TLVILPDD182 pKa = 3.55GCGTILHH189 pKa = 7.04AFYY192 pKa = 10.81CVLHH196 pKa = 6.74PRR198 pKa = 11.84TQQNCAGEE206 pKa = 4.56TNFKK210 pKa = 10.74SLSLWDD216 pKa = 4.4TPASDD221 pKa = 4.53CVSGSYY227 pKa = 9.41NQEE230 pKa = 3.54ATLGAFKK237 pKa = 10.95VYY239 pKa = 10.44FDD241 pKa = 6.08LINCTFRR248 pKa = 11.84YY249 pKa = 10.19NYY251 pKa = 9.77TITEE255 pKa = 4.22DD256 pKa = 4.48EE257 pKa = 3.97NAEE260 pKa = 4.14WFGITQDD267 pKa = 3.47TQGVHH272 pKa = 6.7LYY274 pKa = 10.45SSRR277 pKa = 11.84KK278 pKa = 9.46EE279 pKa = 3.81NVFRR283 pKa = 11.84NNMFHH288 pKa = 7.46FATLPVYY295 pKa = 10.33QKK297 pKa = 10.09ILYY300 pKa = 7.25YY301 pKa = 9.63TVIPRR306 pKa = 11.84SIRR309 pKa = 11.84SPFNDD314 pKa = 2.95RR315 pKa = 11.84KK316 pKa = 10.37AWAAFYY322 pKa = 9.99IYY324 pKa = 10.33KK325 pKa = 10.23LHH327 pKa = 7.02PLTYY331 pKa = 10.54LLNFDD336 pKa = 3.62VEE338 pKa = 4.81GYY340 pKa = 7.31ITKK343 pKa = 10.51AVDD346 pKa = 3.43CGYY349 pKa = 11.23DD350 pKa = 4.06DD351 pKa = 5.81LAQLQCSYY359 pKa = 11.37EE360 pKa = 4.24SFEE363 pKa = 4.45VEE365 pKa = 3.78TGVYY369 pKa = 9.84SVSSFEE375 pKa = 3.78ASPRR379 pKa = 11.84GEE381 pKa = 4.58FIEE384 pKa = 4.21QATTQEE390 pKa = 4.29CDD392 pKa = 3.74FTPMLTGTPPPIYY405 pKa = 9.95NFKK408 pKa = 10.75RR409 pKa = 11.84LVFTNCNYY417 pKa = 10.82NLTKK421 pKa = 10.58LLSLFQVSEE430 pKa = 4.71FSCHH434 pKa = 4.9QVSPSSLATGCYY446 pKa = 9.95SSLTVDD452 pKa = 3.43YY453 pKa = 9.87FAYY456 pKa = 8.82STDD459 pKa = 3.07MSSYY463 pKa = 9.68LQPGSAGAIVQFNYY477 pKa = 10.33KK478 pKa = 9.78QDD480 pKa = 3.66FSNPTCRR487 pKa = 11.84VLATVPQNLTTITKK501 pKa = 8.73PSNYY505 pKa = 10.1AYY507 pKa = 9.51LTEE510 pKa = 4.67CYY512 pKa = 7.89KK513 pKa = 10.6TSAYY517 pKa = 9.25GKK519 pKa = 9.32NYY521 pKa = 10.18LYY523 pKa = 10.26NAPGAYY529 pKa = 8.3TPCLSLASRR538 pKa = 11.84GFSTKK543 pKa = 10.14YY544 pKa = 9.57QSHH547 pKa = 6.82SDD549 pKa = 3.76GEE551 pKa = 4.52LTTTGYY557 pKa = 10.29IYY559 pKa = 10.33PVTGNLQMAFIISVQYY575 pKa = 9.07GTDD578 pKa = 3.55TNSVCPMQALRR589 pKa = 11.84NDD591 pKa = 3.48TSIEE595 pKa = 4.13DD596 pKa = 3.73KK597 pKa = 11.26LDD599 pKa = 3.35VCVEE603 pKa = 3.97YY604 pKa = 10.66SLHH607 pKa = 6.69GITGRR612 pKa = 11.84GVFHH616 pKa = 7.07NCTSVGLRR624 pKa = 11.84NQRR627 pKa = 11.84FVYY630 pKa = 9.22DD631 pKa = 3.57TFDD634 pKa = 3.52NLVGYY639 pKa = 10.57HH640 pKa = 6.66SDD642 pKa = 3.15NGNYY646 pKa = 9.55YY647 pKa = 9.96CVRR650 pKa = 11.84PCVSVPVSVIYY661 pKa = 10.76DD662 pKa = 3.52KK663 pKa = 11.4ASNSHH668 pKa = 5.69ATLFGSVACSHH679 pKa = 5.84VTTMMSQFSRR689 pKa = 11.84MTKK692 pKa = 9.37TNLLARR698 pKa = 11.84TTPGPLQTTVGCAMGFINSSMVVDD722 pKa = 4.66EE723 pKa = 4.72CQLPLGQSLCAIPPTTSSRR742 pKa = 11.84VRR744 pKa = 11.84RR745 pKa = 11.84ATSGASDD752 pKa = 3.62VFQIATLNFTSPLTLAPINSTGFVVAVPTNFTFGVTQEE790 pKa = 4.56FIEE793 pKa = 4.32TTIQKK798 pKa = 8.34ITVDD802 pKa = 3.89CKK804 pKa = 10.81QYY806 pKa = 10.53VCNGFKK812 pKa = 10.45KK813 pKa = 10.92CEE815 pKa = 4.15DD816 pKa = 3.47LLKK819 pKa = 10.63EE820 pKa = 4.03YY821 pKa = 10.97GQFCSKK827 pKa = 10.24INQALHH833 pKa = 5.92GANLRR838 pKa = 11.84QDD840 pKa = 3.5EE841 pKa = 4.74SIANLFSSIKK851 pKa = 8.36TQNTQPLQAGLNGDD865 pKa = 4.19FNLTMLQIPQVTTGEE880 pKa = 3.87RR881 pKa = 11.84KK882 pKa = 9.32YY883 pKa = 10.94RR884 pKa = 11.84STIEE888 pKa = 4.38DD889 pKa = 3.78LLFNKK894 pKa = 7.82VTIADD899 pKa = 3.7PGYY902 pKa = 7.86MQGYY906 pKa = 8.62DD907 pKa = 2.97EE908 pKa = 5.92CMQQGPQSARR918 pKa = 11.84DD919 pKa = 4.21LICAQYY925 pKa = 9.2VAGYY929 pKa = 9.86KK930 pKa = 9.71VLPPLYY936 pKa = 9.9DD937 pKa = 3.96PYY939 pKa = 10.78MEE941 pKa = 4.81AAYY944 pKa = 9.18TSSLLGSIAGASWTAGLSSFAAIPFAQSIFYY975 pKa = 10.26RR976 pKa = 11.84LNGVGITQQVLSEE989 pKa = 4.05NQKK992 pKa = 10.46IIANKK997 pKa = 9.42FNQALGAMQTGFTTTNLAFNKK1018 pKa = 9.95VQDD1021 pKa = 3.97AVNANAMALSKK1032 pKa = 10.7LAAEE1036 pKa = 4.98LSNTFGAISSSISDD1050 pKa = 3.1ILARR1054 pKa = 11.84LDD1056 pKa = 3.62TVEE1059 pKa = 5.08QEE1061 pKa = 4.13AQIDD1065 pKa = 3.71RR1066 pKa = 11.84LINGRR1071 pKa = 11.84LTSLNAFVAQQLVRR1085 pKa = 11.84TEE1087 pKa = 3.65AAARR1091 pKa = 11.84SAQLAQDD1098 pKa = 3.82KK1099 pKa = 11.07VNEE1102 pKa = 4.39CVKK1105 pKa = 10.74SQSKK1109 pKa = 10.87RR1110 pKa = 11.84NGFCGTGTHH1119 pKa = 5.85IVSFAINAPNGLYY1132 pKa = 10.03FFHH1135 pKa = 7.75VGYY1138 pKa = 10.58QPTSHH1143 pKa = 6.33VNATAAYY1150 pKa = 8.36GLCNTEE1156 pKa = 4.45NPQKK1160 pKa = 10.6CIAPIDD1166 pKa = 4.22GYY1168 pKa = 11.21FVLNQTTSTVADD1180 pKa = 4.45SDD1182 pKa = 3.99QQWYY1186 pKa = 7.3YY1187 pKa = 9.08TGSSFFHH1194 pKa = 6.96PEE1196 pKa = 4.31PITEE1200 pKa = 4.24ANSKK1204 pKa = 8.77YY1205 pKa = 11.08VSMDD1209 pKa = 3.11VKK1211 pKa = 11.03FEE1213 pKa = 4.13NLTNRR1218 pKa = 11.84LPPPLLSNSTDD1229 pKa = 3.39LDD1231 pKa = 3.81FKK1233 pKa = 11.54EE1234 pKa = 4.12EE1235 pKa = 4.03LEE1237 pKa = 4.22EE1238 pKa = 4.02FFKK1241 pKa = 11.25NVSSQGPNFQEE1252 pKa = 3.42ISKK1255 pKa = 10.78INTTLLNLNTEE1266 pKa = 4.26LMVLSEE1272 pKa = 4.38VVKK1275 pKa = 10.44QLNEE1279 pKa = 3.95SYY1281 pKa = 10.82IDD1283 pKa = 3.87LKK1285 pKa = 11.29EE1286 pKa = 4.03LGNYY1290 pKa = 7.36TFYY1293 pKa = 11.09QKK1295 pKa = 10.17WPWYY1299 pKa = 8.52IWLGFIAGLVALALCVFFILCCTGCGTSCLGKK1331 pKa = 10.25LKK1333 pKa = 10.69CNRR1336 pKa = 11.84CCDD1339 pKa = 3.46SYY1341 pKa = 12.06DD1342 pKa = 3.43EE1343 pKa = 4.74YY1344 pKa = 10.97EE1345 pKa = 4.0VEE1347 pKa = 4.86KK1348 pKa = 10.84IHH1350 pKa = 6.28VHH1352 pKa = 5.56
Molecular weight: 149.7 kDa
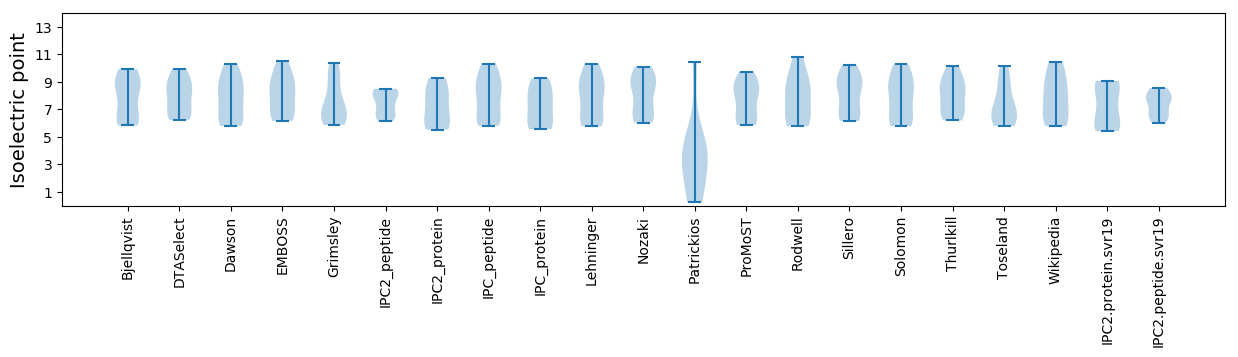
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|P0C6T5|R1A_BCHK5 Replicase polyprotein 1a OS=Bat coronavirus HKU5 OX=694008 GN=1a PE=3 SV=1
MM1 pKa = 7.4ATPAPPRR8 pKa = 11.84AVVFANDD15 pKa = 3.47NEE17 pKa = 4.68TPTNSQRR24 pKa = 11.84SGRR27 pKa = 11.84PRR29 pKa = 11.84TKK31 pKa = 10.13PRR33 pKa = 11.84PAPNTTVSWFTGLTQHH49 pKa = 6.72GKK51 pKa = 9.68QPLAFPPGQGVPLNANSTPAQNAGYY76 pKa = 9.33WRR78 pKa = 11.84RR79 pKa = 11.84QDD81 pKa = 3.96RR82 pKa = 11.84KK83 pKa = 10.03INTGNGTKK91 pKa = 10.09PLAPRR96 pKa = 11.84WYY98 pKa = 9.82FYY100 pKa = 10.45YY101 pKa = 10.42TGTGPEE107 pKa = 3.84ANLPFRR113 pKa = 11.84SVKK116 pKa = 10.66DD117 pKa = 3.63GIIWVHH123 pKa = 5.87EE124 pKa = 4.29NGATDD129 pKa = 3.49APSVFGTRR137 pKa = 11.84NPANDD142 pKa = 3.9PAIVTQFAPGTTLPKK157 pKa = 10.23NFHH160 pKa = 6.56IEE162 pKa = 4.18GTGGNSQSSSRR173 pKa = 11.84ASSRR177 pKa = 11.84SSSRR181 pKa = 11.84SSSRR185 pKa = 11.84NGRR188 pKa = 11.84SNNSSRR194 pKa = 11.84NASPAPHH201 pKa = 6.41GVGDD205 pKa = 3.85VVGAGTLSVLLDD217 pKa = 3.75LQKK220 pKa = 11.11RR221 pKa = 11.84LADD224 pKa = 3.87LEE226 pKa = 4.35AGKK229 pKa = 10.82GNKK232 pKa = 7.63QPKK235 pKa = 10.13VITKK239 pKa = 10.13KK240 pKa = 10.37DD241 pKa = 3.21AQAAKK246 pKa = 10.25QKK248 pKa = 9.14MRR250 pKa = 11.84HH251 pKa = 5.5KK252 pKa = 10.47RR253 pKa = 11.84VATKK257 pKa = 10.47GYY259 pKa = 10.6NVVQAFGMRR268 pKa = 11.84GPGPLQSNFGDD279 pKa = 3.44MQYY282 pKa = 11.39NKK284 pKa = 10.42LGTEE288 pKa = 4.21DD289 pKa = 4.05PRR291 pKa = 11.84WPQIAEE297 pKa = 4.2LAPSASAFMSTSQFKK312 pKa = 8.73VTHH315 pKa = 5.74QSNDD319 pKa = 3.34EE320 pKa = 4.17NGEE323 pKa = 3.87PVYY326 pKa = 10.52FLSYY330 pKa = 10.63SGAIKK335 pKa = 10.63LDD337 pKa = 3.71PKK339 pKa = 10.57NPNYY343 pKa = 9.8NKK345 pKa = 9.83WMEE348 pKa = 4.15ILDD351 pKa = 4.08ANIDD355 pKa = 3.74AYY357 pKa = 10.21KK358 pKa = 10.59SFPKK362 pKa = 10.11KK363 pKa = 9.37EE364 pKa = 4.04RR365 pKa = 11.84KK366 pKa = 9.04QKK368 pKa = 10.65PSGDD372 pKa = 3.97DD373 pKa = 3.06AATAPATSQMEE384 pKa = 4.7DD385 pKa = 3.62VPEE388 pKa = 4.56LPPKK392 pKa = 7.94QQRR395 pKa = 11.84KK396 pKa = 8.62KK397 pKa = 10.5RR398 pKa = 11.84VVQGSIPQRR407 pKa = 11.84SAGVPSFEE415 pKa = 5.04DD416 pKa = 3.28VVDD419 pKa = 5.41AIFPDD424 pKa = 4.2SEE426 pKa = 4.17AA427 pKa = 4.08
MM1 pKa = 7.4ATPAPPRR8 pKa = 11.84AVVFANDD15 pKa = 3.47NEE17 pKa = 4.68TPTNSQRR24 pKa = 11.84SGRR27 pKa = 11.84PRR29 pKa = 11.84TKK31 pKa = 10.13PRR33 pKa = 11.84PAPNTTVSWFTGLTQHH49 pKa = 6.72GKK51 pKa = 9.68QPLAFPPGQGVPLNANSTPAQNAGYY76 pKa = 9.33WRR78 pKa = 11.84RR79 pKa = 11.84QDD81 pKa = 3.96RR82 pKa = 11.84KK83 pKa = 10.03INTGNGTKK91 pKa = 10.09PLAPRR96 pKa = 11.84WYY98 pKa = 9.82FYY100 pKa = 10.45YY101 pKa = 10.42TGTGPEE107 pKa = 3.84ANLPFRR113 pKa = 11.84SVKK116 pKa = 10.66DD117 pKa = 3.63GIIWVHH123 pKa = 5.87EE124 pKa = 4.29NGATDD129 pKa = 3.49APSVFGTRR137 pKa = 11.84NPANDD142 pKa = 3.9PAIVTQFAPGTTLPKK157 pKa = 10.23NFHH160 pKa = 6.56IEE162 pKa = 4.18GTGGNSQSSSRR173 pKa = 11.84ASSRR177 pKa = 11.84SSSRR181 pKa = 11.84SSSRR185 pKa = 11.84NGRR188 pKa = 11.84SNNSSRR194 pKa = 11.84NASPAPHH201 pKa = 6.41GVGDD205 pKa = 3.85VVGAGTLSVLLDD217 pKa = 3.75LQKK220 pKa = 11.11RR221 pKa = 11.84LADD224 pKa = 3.87LEE226 pKa = 4.35AGKK229 pKa = 10.82GNKK232 pKa = 7.63QPKK235 pKa = 10.13VITKK239 pKa = 10.13KK240 pKa = 10.37DD241 pKa = 3.21AQAAKK246 pKa = 10.25QKK248 pKa = 9.14MRR250 pKa = 11.84HH251 pKa = 5.5KK252 pKa = 10.47RR253 pKa = 11.84VATKK257 pKa = 10.47GYY259 pKa = 10.6NVVQAFGMRR268 pKa = 11.84GPGPLQSNFGDD279 pKa = 3.44MQYY282 pKa = 11.39NKK284 pKa = 10.42LGTEE288 pKa = 4.21DD289 pKa = 4.05PRR291 pKa = 11.84WPQIAEE297 pKa = 4.2LAPSASAFMSTSQFKK312 pKa = 8.73VTHH315 pKa = 5.74QSNDD319 pKa = 3.34EE320 pKa = 4.17NGEE323 pKa = 3.87PVYY326 pKa = 10.52FLSYY330 pKa = 10.63SGAIKK335 pKa = 10.63LDD337 pKa = 3.71PKK339 pKa = 10.57NPNYY343 pKa = 9.8NKK345 pKa = 9.83WMEE348 pKa = 4.15ILDD351 pKa = 4.08ANIDD355 pKa = 3.74AYY357 pKa = 10.21KK358 pKa = 10.59SFPKK362 pKa = 10.11KK363 pKa = 9.37EE364 pKa = 4.04RR365 pKa = 11.84KK366 pKa = 9.04QKK368 pKa = 10.65PSGDD372 pKa = 3.97DD373 pKa = 3.06AATAPATSQMEE384 pKa = 4.7DD385 pKa = 3.62VPEE388 pKa = 4.56LPPKK392 pKa = 7.94QQRR395 pKa = 11.84KK396 pKa = 8.62KK397 pKa = 10.5RR398 pKa = 11.84VVQGSIPQRR407 pKa = 11.84SAGVPSFEE415 pKa = 5.04DD416 pKa = 3.28VVDD419 pKa = 5.41AIFPDD424 pKa = 4.2SEE426 pKa = 4.17AA427 pKa = 4.08
Molecular weight: 46.29 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
14463 |
82 |
7182 |
1446.3 |
160.66 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.585 ± 0.214 | 3.049 ± 0.264 |
5.352 ± 0.26 | 4.066 ± 0.135 |
4.653 ± 0.169 | 5.905 ± 0.177 |
2.026 ± 0.105 | 4.556 ± 0.157 |
5.317 ± 0.27 | 9.279 ± 0.389 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.171 ± 0.109 | 5.22 ± 0.194 |
4.107 ± 0.504 | 3.423 ± 0.389 |
3.699 ± 0.322 | 7.488 ± 0.27 |
6.873 ± 0.295 | 9.196 ± 0.585 |
1.203 ± 0.097 | 4.826 ± 0.235 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
