
Halogranum gelatinilyticum
Taxonomy: cellular organisms; Archaea; Euryarchaeota; Stenosarchaea group; Halobacteria; Haloferacales; Haloferacaceae; Halogranum
Average proteome isoelectric point is 4.93
Get precalculated fractions of proteins
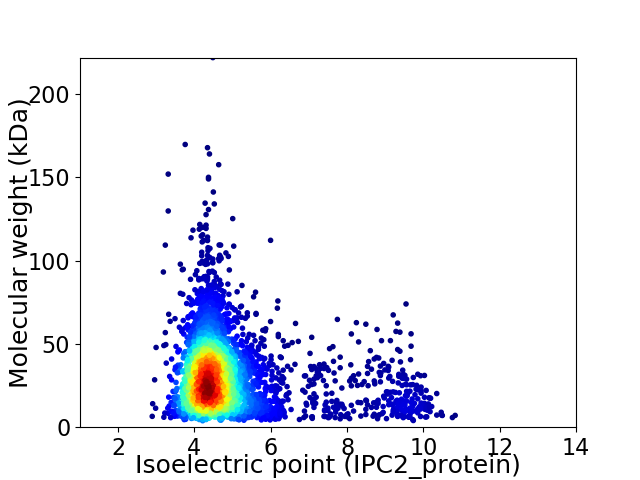
Virtual 2D-PAGE plot for 3752 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
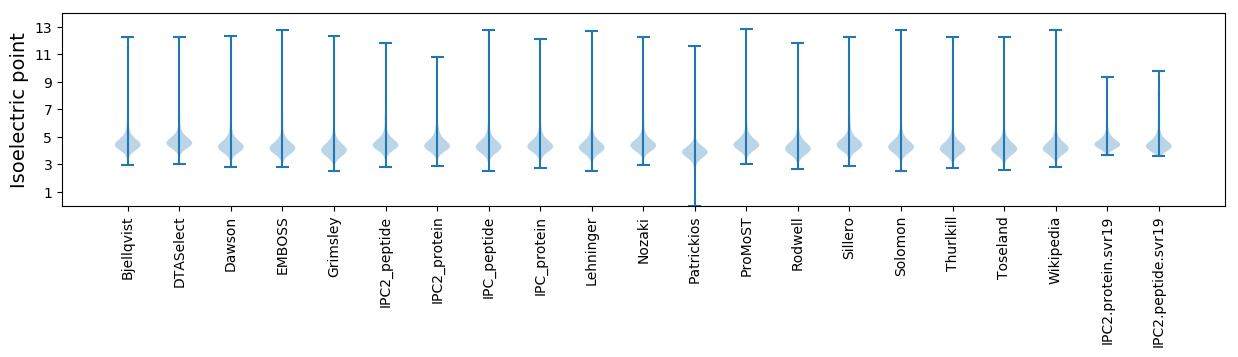
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1G9TGR2|A0A1G9TGR2_9EURY Uncharacterized protein OS=Halogranum gelatinilyticum OX=660521 GN=SAMN04487949_1768 PE=4 SV=1
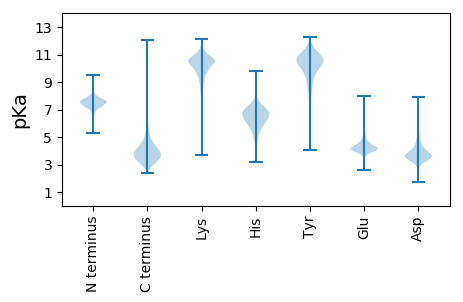
MM1 pKa = 6.93TQDD4 pKa = 3.46GTPDD8 pKa = 3.17LNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84FLQATGAVGLAGALGGVASATPGRR37 pKa = 11.84SPGPKK42 pKa = 8.82EE43 pKa = 4.72DD44 pKa = 4.33EE45 pKa = 4.17ILVGVAQGDD54 pKa = 3.95DD55 pKa = 3.63SPADD59 pKa = 3.66VVKK62 pKa = 10.74KK63 pKa = 10.36AVPGKK68 pKa = 10.58ARR70 pKa = 11.84IVHH73 pKa = 5.44EE74 pKa = 4.7NKK76 pKa = 8.83TLSYY80 pKa = 10.49VAVKK84 pKa = 10.25FPSQAADD91 pKa = 3.24VARR94 pKa = 11.84EE95 pKa = 3.92NFIEE99 pKa = 4.7AVTKK103 pKa = 10.47NDD105 pKa = 3.13RR106 pKa = 11.84VKK108 pKa = 10.83YY109 pKa = 9.58AEE111 pKa = 4.2KK112 pKa = 10.57NATLSTSLSPNDD124 pKa = 3.57PRR126 pKa = 11.84YY127 pKa = 10.03GDD129 pKa = 3.3QYY131 pKa = 11.1APQMVNADD139 pKa = 4.21TAWDD143 pKa = 3.85TTLGSSSVTIAVVDD157 pKa = 3.4TGVQYY162 pKa = 10.47DD163 pKa = 4.0HH164 pKa = 7.89PDD166 pKa = 3.15LAGSFGSNPGYY177 pKa = 10.95DD178 pKa = 3.63FVDD181 pKa = 4.6DD182 pKa = 5.92DD183 pKa = 4.79GDD185 pKa = 4.03PYY187 pKa = 11.25PDD189 pKa = 3.39VLADD193 pKa = 4.19EE194 pKa = 4.48YY195 pKa = 11.08HH196 pKa = 5.62GTHH199 pKa = 5.66VAGIAGGVTDD209 pKa = 4.69NGTGTSGISNSTLISGRR226 pKa = 11.84ALDD229 pKa = 3.64EE230 pKa = 4.35SGRR233 pKa = 11.84GSTADD238 pKa = 2.99IADD241 pKa = 3.79AVEE244 pKa = 3.79WAADD248 pKa = 3.45QGADD252 pKa = 4.73VINLSLGGGGYY263 pKa = 9.07TSTMKK268 pKa = 10.69NAVSYY273 pKa = 8.31ATNNGALVVAAAGNNGTSSVSYY295 pKa = 8.65PAAYY299 pKa = 9.87SEE301 pKa = 4.4CLAVSSLDD309 pKa = 3.55PDD311 pKa = 4.01GSLASYY317 pKa = 9.41SQYY320 pKa = 11.02GSSIEE325 pKa = 3.94LAAPGSNVLATLPTDD340 pKa = 4.4DD341 pKa = 4.44YY342 pKa = 11.7GEE344 pKa = 4.35LSGTSMATPVVAGVAGLTLAQWDD367 pKa = 3.91VDD369 pKa = 3.72NVTLRR374 pKa = 11.84DD375 pKa = 3.81HH376 pKa = 7.11LKK378 pKa = 8.74NTAVDD383 pKa = 3.33IGLSSSQQGSGRR395 pKa = 11.84VDD397 pKa = 2.82AANAVEE403 pKa = 4.18TDD405 pKa = 3.59PANGGGGDD413 pKa = 4.04GGDD416 pKa = 3.92GGDD419 pKa = 4.46DD420 pKa = 3.76GGSGGSTSASVTDD433 pKa = 3.99SLTNSADD440 pKa = 4.01SKK442 pKa = 9.22CWTYY446 pKa = 11.48AFEE449 pKa = 4.3YY450 pKa = 10.05SNPSTVVISLDD461 pKa = 3.48GPSYY465 pKa = 11.73ADD467 pKa = 3.37FDD469 pKa = 5.07LYY471 pKa = 11.57ANDD474 pKa = 4.17GTGTCPSTSSYY485 pKa = 11.27DD486 pKa = 3.55YY487 pKa = 10.99RR488 pKa = 11.84SWTTTSQEE496 pKa = 4.7SITIDD501 pKa = 4.02NPDD504 pKa = 3.15TSADD508 pKa = 3.55LSILVDD514 pKa = 3.83SYY516 pKa = 11.69SGSGSYY522 pKa = 8.54TLEE525 pKa = 3.68IVEE528 pKa = 4.42YY529 pKa = 9.92EE530 pKa = 4.0
MM1 pKa = 6.93TQDD4 pKa = 3.46GTPDD8 pKa = 3.17LNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84FLQATGAVGLAGALGGVASATPGRR37 pKa = 11.84SPGPKK42 pKa = 8.82EE43 pKa = 4.72DD44 pKa = 4.33EE45 pKa = 4.17ILVGVAQGDD54 pKa = 3.95DD55 pKa = 3.63SPADD59 pKa = 3.66VVKK62 pKa = 10.74KK63 pKa = 10.36AVPGKK68 pKa = 10.58ARR70 pKa = 11.84IVHH73 pKa = 5.44EE74 pKa = 4.7NKK76 pKa = 8.83TLSYY80 pKa = 10.49VAVKK84 pKa = 10.25FPSQAADD91 pKa = 3.24VARR94 pKa = 11.84EE95 pKa = 3.92NFIEE99 pKa = 4.7AVTKK103 pKa = 10.47NDD105 pKa = 3.13RR106 pKa = 11.84VKK108 pKa = 10.83YY109 pKa = 9.58AEE111 pKa = 4.2KK112 pKa = 10.57NATLSTSLSPNDD124 pKa = 3.57PRR126 pKa = 11.84YY127 pKa = 10.03GDD129 pKa = 3.3QYY131 pKa = 11.1APQMVNADD139 pKa = 4.21TAWDD143 pKa = 3.85TTLGSSSVTIAVVDD157 pKa = 3.4TGVQYY162 pKa = 10.47DD163 pKa = 4.0HH164 pKa = 7.89PDD166 pKa = 3.15LAGSFGSNPGYY177 pKa = 10.95DD178 pKa = 3.63FVDD181 pKa = 4.6DD182 pKa = 5.92DD183 pKa = 4.79GDD185 pKa = 4.03PYY187 pKa = 11.25PDD189 pKa = 3.39VLADD193 pKa = 4.19EE194 pKa = 4.48YY195 pKa = 11.08HH196 pKa = 5.62GTHH199 pKa = 5.66VAGIAGGVTDD209 pKa = 4.69NGTGTSGISNSTLISGRR226 pKa = 11.84ALDD229 pKa = 3.64EE230 pKa = 4.35SGRR233 pKa = 11.84GSTADD238 pKa = 2.99IADD241 pKa = 3.79AVEE244 pKa = 3.79WAADD248 pKa = 3.45QGADD252 pKa = 4.73VINLSLGGGGYY263 pKa = 9.07TSTMKK268 pKa = 10.69NAVSYY273 pKa = 8.31ATNNGALVVAAAGNNGTSSVSYY295 pKa = 8.65PAAYY299 pKa = 9.87SEE301 pKa = 4.4CLAVSSLDD309 pKa = 3.55PDD311 pKa = 4.01GSLASYY317 pKa = 9.41SQYY320 pKa = 11.02GSSIEE325 pKa = 3.94LAAPGSNVLATLPTDD340 pKa = 4.4DD341 pKa = 4.44YY342 pKa = 11.7GEE344 pKa = 4.35LSGTSMATPVVAGVAGLTLAQWDD367 pKa = 3.91VDD369 pKa = 3.72NVTLRR374 pKa = 11.84DD375 pKa = 3.81HH376 pKa = 7.11LKK378 pKa = 8.74NTAVDD383 pKa = 3.33IGLSSSQQGSGRR395 pKa = 11.84VDD397 pKa = 2.82AANAVEE403 pKa = 4.18TDD405 pKa = 3.59PANGGGGDD413 pKa = 4.04GGDD416 pKa = 3.92GGDD419 pKa = 4.46DD420 pKa = 3.76GGSGGSTSASVTDD433 pKa = 3.99SLTNSADD440 pKa = 4.01SKK442 pKa = 9.22CWTYY446 pKa = 11.48AFEE449 pKa = 4.3YY450 pKa = 10.05SNPSTVVISLDD461 pKa = 3.48GPSYY465 pKa = 11.73ADD467 pKa = 3.37FDD469 pKa = 5.07LYY471 pKa = 11.57ANDD474 pKa = 4.17GTGTCPSTSSYY485 pKa = 11.27DD486 pKa = 3.55YY487 pKa = 10.99RR488 pKa = 11.84SWTTTSQEE496 pKa = 4.7SITIDD501 pKa = 4.02NPDD504 pKa = 3.15TSADD508 pKa = 3.55LSILVDD514 pKa = 3.83SYY516 pKa = 11.69SGSGSYY522 pKa = 8.54TLEE525 pKa = 3.68IVEE528 pKa = 4.42YY529 pKa = 9.92EE530 pKa = 4.0
Molecular weight: 54.07 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1G9ZGJ2|A0A1G9ZGJ2_9EURY Thiamine transport system substrate-binding protein OS=Halogranum gelatinilyticum OX=660521 GN=SAMN04487949_3629 PE=4 SV=1
MM1 pKa = 7.52SPLGAAPLLAPEE13 pKa = 5.43KK14 pKa = 10.34IPSRR18 pKa = 11.84SLRR21 pKa = 11.84SLPTASPPSILVGFWVEE38 pKa = 3.92HH39 pKa = 5.91PRR41 pKa = 11.84FSVEE45 pKa = 3.19KK46 pKa = 10.49RR47 pKa = 11.84MTLRR51 pKa = 11.84CSTHH55 pKa = 5.38FLEE58 pKa = 5.13MVNVPPKK65 pKa = 10.32FRR67 pKa = 11.84IGGTPRR73 pKa = 11.84SLAILWLEE81 pKa = 4.13ANRR84 pKa = 11.84KK85 pKa = 9.02NGWNTPCFTVCRR97 pKa = 11.84YY98 pKa = 9.22PISLPSLPPLIQYY111 pKa = 9.68HH112 pKa = 6.52DD113 pKa = 3.93PEE115 pKa = 4.53KK116 pKa = 10.92NRR118 pKa = 11.84YY119 pKa = 5.4RR120 pKa = 11.84TQRR123 pKa = 11.84KK124 pKa = 8.83RR125 pKa = 11.84EE126 pKa = 4.05QVPHH130 pKa = 5.64QAVADD135 pKa = 4.71RR136 pKa = 11.84SQIQCDD142 pKa = 3.55FRR144 pKa = 11.84AFVRR148 pKa = 11.84TNSGSQGRR156 pKa = 11.84GHH158 pKa = 6.18VALPTLHH165 pKa = 6.74PRR167 pKa = 11.84ITAPLNRR174 pKa = 11.84VSSQLLPCANFLSRR188 pKa = 11.84YY189 pKa = 8.79VIYY192 pKa = 10.64KK193 pKa = 10.22RR194 pKa = 11.84ALLRR198 pKa = 11.84NTGDD202 pKa = 3.3SDD204 pKa = 3.8RR205 pKa = 11.84QTSEE209 pKa = 4.02LALEE213 pKa = 4.34TTLLITPIITSRR225 pKa = 11.84LL226 pKa = 3.49
MM1 pKa = 7.52SPLGAAPLLAPEE13 pKa = 5.43KK14 pKa = 10.34IPSRR18 pKa = 11.84SLRR21 pKa = 11.84SLPTASPPSILVGFWVEE38 pKa = 3.92HH39 pKa = 5.91PRR41 pKa = 11.84FSVEE45 pKa = 3.19KK46 pKa = 10.49RR47 pKa = 11.84MTLRR51 pKa = 11.84CSTHH55 pKa = 5.38FLEE58 pKa = 5.13MVNVPPKK65 pKa = 10.32FRR67 pKa = 11.84IGGTPRR73 pKa = 11.84SLAILWLEE81 pKa = 4.13ANRR84 pKa = 11.84KK85 pKa = 9.02NGWNTPCFTVCRR97 pKa = 11.84YY98 pKa = 9.22PISLPSLPPLIQYY111 pKa = 9.68HH112 pKa = 6.52DD113 pKa = 3.93PEE115 pKa = 4.53KK116 pKa = 10.92NRR118 pKa = 11.84YY119 pKa = 5.4RR120 pKa = 11.84TQRR123 pKa = 11.84KK124 pKa = 8.83RR125 pKa = 11.84EE126 pKa = 4.05QVPHH130 pKa = 5.64QAVADD135 pKa = 4.71RR136 pKa = 11.84SQIQCDD142 pKa = 3.55FRR144 pKa = 11.84AFVRR148 pKa = 11.84TNSGSQGRR156 pKa = 11.84GHH158 pKa = 6.18VALPTLHH165 pKa = 6.74PRR167 pKa = 11.84ITAPLNRR174 pKa = 11.84VSSQLLPCANFLSRR188 pKa = 11.84YY189 pKa = 8.79VIYY192 pKa = 10.64KK193 pKa = 10.22RR194 pKa = 11.84ALLRR198 pKa = 11.84NTGDD202 pKa = 3.3SDD204 pKa = 3.8RR205 pKa = 11.84QTSEE209 pKa = 4.02LALEE213 pKa = 4.34TTLLITPIITSRR225 pKa = 11.84LL226 pKa = 3.49
Molecular weight: 25.57 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1098223 |
39 |
1990 |
292.7 |
31.78 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.67 ± 0.058 | 0.682 ± 0.013 |
8.315 ± 0.048 | 8.046 ± 0.052 |
3.449 ± 0.026 | 8.567 ± 0.039 |
1.983 ± 0.021 | 3.644 ± 0.034 |
2.057 ± 0.03 | 9.226 ± 0.053 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.753 ± 0.019 | 2.425 ± 0.025 |
4.561 ± 0.026 | 2.641 ± 0.024 |
6.334 ± 0.033 | 5.624 ± 0.032 |
6.605 ± 0.028 | 9.541 ± 0.042 |
1.155 ± 0.016 | 2.722 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
