
Flavobacterium sp. 123
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Flavobacterium; unclassified Flavobacterium
Average proteome isoelectric point is 6.66
Get precalculated fractions of proteins
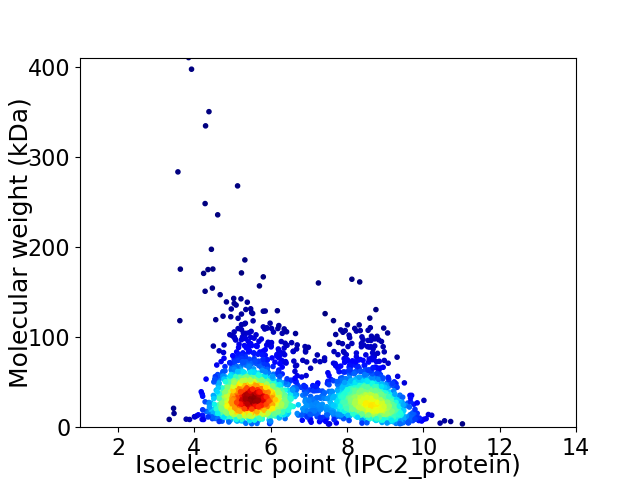
Virtual 2D-PAGE plot for 2513 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A495SLF6|A0A495SLF6_9FLAO Acetyl-CoA acyltransferase OS=Flavobacterium sp. 123 OX=2135627 GN=C8C88_2168 PE=3 SV=1
MM1 pKa = 7.56KK2 pKa = 10.42LKK4 pKa = 10.74NYY6 pKa = 10.18LPNQLIKK13 pKa = 10.7CNQEE17 pKa = 3.56STSYY21 pKa = 10.09SKK23 pKa = 11.01KK24 pKa = 10.05VFLFFAALFLTVASSYY40 pKa = 11.23SLEE43 pKa = 4.1AQNVSLTFSKK53 pKa = 10.84GYY55 pKa = 10.56LGTQGSNTNQANSINNLSNIGINRR79 pKa = 11.84ISFSQAYY86 pKa = 9.36AGAFGGTQGNDD97 pKa = 2.82LSGNLTFYY105 pKa = 10.71LANGTSFSLTGALNWRR121 pKa = 11.84EE122 pKa = 4.12TSGSTVEE129 pKa = 3.95VFGFIFDD136 pKa = 3.62SGQNHH141 pKa = 7.35TITYY145 pKa = 10.77GLGQTYY151 pKa = 10.54NIVGGSTPNTSTTLGLKK168 pKa = 10.03AYY170 pKa = 9.45ASSFVFVNGEE180 pKa = 3.74NRR182 pKa = 11.84AGNAATNGLLDD193 pKa = 4.06ALNAEE198 pKa = 5.13LLSSPQPSTVTLSTANVTEE217 pKa = 4.18GQNLVYY223 pKa = 10.74NVTLSSATTVGNPQTFVFTYY243 pKa = 10.55GGTSTNGSDD252 pKa = 3.65YY253 pKa = 11.27NSTFTFSNGVIDD265 pKa = 5.01NGDD268 pKa = 3.38GTITVPGGVSSFTITASTTDD288 pKa = 3.44DD289 pKa = 3.38SSIEE293 pKa = 4.1STEE296 pKa = 3.95TLILNIGSKK305 pKa = 10.2SDD307 pKa = 3.3SGNILDD313 pKa = 4.5NDD315 pKa = 4.22SVALTASTSQTNLFCFGGSNGTASVVVSGGTAPYY349 pKa = 9.62SYY351 pKa = 10.44SWSPSGGTWATASGLTAGNYY371 pKa = 7.39TVTITDD377 pKa = 4.36DD378 pKa = 3.47NLATITKK385 pKa = 9.28NFTITEE391 pKa = 4.32PSAITASASQTNVSCNGGSNGTASVTPSGGAGSYY425 pKa = 9.0TYY427 pKa = 10.52SWSPSGGTGATASGLSAGSYY447 pKa = 8.99TVTIEE452 pKa = 4.44DD453 pKa = 4.54ANGCSATRR461 pKa = 11.84NFTITEE467 pKa = 4.26PSAIAASASQTNIACNGGATGSASVTPSGGAGSYY501 pKa = 9.0TYY503 pKa = 10.52SWSPSGGTGATASGLTAGNYY523 pKa = 7.39TVTITDD529 pKa = 4.04ANLCTATRR537 pKa = 11.84NFTITEE543 pKa = 4.3PSAITATTSQTNTSCNGSTDD563 pKa = 4.29GIASVIASGGTGSYY577 pKa = 10.04SYY579 pKa = 10.72SWSPSGGTAATATGLAAGNYY599 pKa = 6.88TCTITDD605 pKa = 4.26LNGCSIDD612 pKa = 3.67KK613 pKa = 10.63NFTIALSSSWNDD625 pKa = 3.15FDD627 pKa = 6.33CDD629 pKa = 3.75GDD631 pKa = 4.08GVTNGTEE638 pKa = 4.25VTDD641 pKa = 3.82GTNPDD646 pKa = 3.49NSCSFTASSITLNASSSWNTADD668 pKa = 4.57CDD670 pKa = 3.83GDD672 pKa = 3.79NVTNGQEE679 pKa = 4.31LLDD682 pKa = 3.99GTDD685 pKa = 3.54PNKK688 pKa = 10.31PDD690 pKa = 3.53TDD692 pKa = 3.91GDD694 pKa = 4.18GVTDD698 pKa = 4.12GKK700 pKa = 11.16EE701 pKa = 4.01KK702 pKa = 10.28TDD704 pKa = 3.48EE705 pKa = 4.09TDD707 pKa = 3.02ATDD710 pKa = 3.36PCKK713 pKa = 10.28FVFASQSVAPSSSWNTADD731 pKa = 4.57CDD733 pKa = 3.83GDD735 pKa = 3.79NVTNGQEE742 pKa = 4.27KK743 pKa = 10.89LDD745 pKa = 3.92GTDD748 pKa = 3.28SNKK751 pKa = 9.89PDD753 pKa = 3.57TDD755 pKa = 3.69GDD757 pKa = 4.18GVTDD761 pKa = 4.15GKK763 pKa = 11.02EE764 pKa = 3.86KK765 pKa = 10.41TDD767 pKa = 3.4ATDD770 pKa = 3.31ATDD773 pKa = 3.32PCKK776 pKa = 10.28FVFASQSVTPSSSWNTADD794 pKa = 5.11CDD796 pKa = 4.05GDD798 pKa = 4.11TITNGQEE805 pKa = 4.16KK806 pKa = 10.68LDD808 pKa = 3.97GTDD811 pKa = 3.21PTKK814 pKa = 10.67PDD816 pKa = 3.39TDD818 pKa = 3.89GDD820 pKa = 4.23GVTDD824 pKa = 4.64DD825 pKa = 5.05KK826 pKa = 11.61EE827 pKa = 4.31KK828 pKa = 10.65TDD830 pKa = 3.74ATDD833 pKa = 3.31ATDD836 pKa = 3.32PCKK839 pKa = 10.28FVFASQSVAPSSSWNTGDD857 pKa = 4.56CDD859 pKa = 4.26GDD861 pKa = 3.69NVTNGQEE868 pKa = 4.31KK869 pKa = 10.86LDD871 pKa = 3.9GTDD874 pKa = 3.66PNVADD879 pKa = 3.92TDD881 pKa = 4.2GDD883 pKa = 4.15GVTDD887 pKa = 4.39DD888 pKa = 4.71KK889 pKa = 11.71EE890 pKa = 4.39KK891 pKa = 10.69TDD893 pKa = 3.48VTDD896 pKa = 4.27ATDD899 pKa = 3.19ACQFILASQSVAPRR913 pKa = 11.84SSWNTADD920 pKa = 4.11CDD922 pKa = 3.74GDD924 pKa = 3.84NVSNGQEE931 pKa = 4.5KK932 pKa = 10.02IDD934 pKa = 3.74GTDD937 pKa = 3.35PLKK940 pKa = 10.77PDD942 pKa = 3.44TDD944 pKa = 4.09GDD946 pKa = 4.28GVTDD950 pKa = 3.83GTEE953 pKa = 4.41KK954 pKa = 10.18IDD956 pKa = 3.47GTNAKK961 pKa = 7.86NTCDD965 pKa = 3.67YY966 pKa = 10.94LAASQTLSPSALWNDD981 pKa = 3.76ADD983 pKa = 4.86CDD985 pKa = 3.67NDD987 pKa = 4.54GIRR990 pKa = 11.84NGDD993 pKa = 3.87DD994 pKa = 3.07NCVFTANANQTDD1006 pKa = 3.75NDD1008 pKa = 3.73NDD1010 pKa = 4.27GLGDD1014 pKa = 3.73TCDD1017 pKa = 5.63DD1018 pKa = 5.02DD1019 pKa = 6.69DD1020 pKa = 6.85DD1021 pKa = 5.98NDD1023 pKa = 5.22GILDD1027 pKa = 3.97TVDD1030 pKa = 3.44NCPLTPNSDD1039 pKa = 3.47QADD1042 pKa = 3.73RR1043 pKa = 11.84DD1044 pKa = 3.76HH1045 pKa = 7.53DD1046 pKa = 4.32GKK1048 pKa = 11.69GDD1050 pKa = 3.66VCDD1053 pKa = 4.54LGLNVSQAITPNGDD1067 pKa = 3.68GINDD1071 pKa = 2.8TWMIYY1076 pKa = 10.4NIEE1079 pKa = 4.17QYY1081 pKa = 10.21PNSTVRR1087 pKa = 11.84VFNRR1091 pKa = 11.84WGSEE1095 pKa = 2.89IFYY1098 pKa = 10.57AHH1100 pKa = 7.43NYY1102 pKa = 8.72RR1103 pKa = 11.84NDD1105 pKa = 3.27WDD1107 pKa = 3.59GHH1109 pKa = 4.92YY1110 pKa = 10.65KK1111 pKa = 10.53NKK1113 pKa = 9.95NQSLPEE1119 pKa = 3.89TSAYY1123 pKa = 10.27YY1124 pKa = 9.61YY1125 pKa = 10.72QIDD1128 pKa = 3.52IEE1130 pKa = 4.82GNGSVDD1136 pKa = 3.77LEE1138 pKa = 3.89GWIYY1142 pKa = 9.21LTKK1145 pKa = 10.84
MM1 pKa = 7.56KK2 pKa = 10.42LKK4 pKa = 10.74NYY6 pKa = 10.18LPNQLIKK13 pKa = 10.7CNQEE17 pKa = 3.56STSYY21 pKa = 10.09SKK23 pKa = 11.01KK24 pKa = 10.05VFLFFAALFLTVASSYY40 pKa = 11.23SLEE43 pKa = 4.1AQNVSLTFSKK53 pKa = 10.84GYY55 pKa = 10.56LGTQGSNTNQANSINNLSNIGINRR79 pKa = 11.84ISFSQAYY86 pKa = 9.36AGAFGGTQGNDD97 pKa = 2.82LSGNLTFYY105 pKa = 10.71LANGTSFSLTGALNWRR121 pKa = 11.84EE122 pKa = 4.12TSGSTVEE129 pKa = 3.95VFGFIFDD136 pKa = 3.62SGQNHH141 pKa = 7.35TITYY145 pKa = 10.77GLGQTYY151 pKa = 10.54NIVGGSTPNTSTTLGLKK168 pKa = 10.03AYY170 pKa = 9.45ASSFVFVNGEE180 pKa = 3.74NRR182 pKa = 11.84AGNAATNGLLDD193 pKa = 4.06ALNAEE198 pKa = 5.13LLSSPQPSTVTLSTANVTEE217 pKa = 4.18GQNLVYY223 pKa = 10.74NVTLSSATTVGNPQTFVFTYY243 pKa = 10.55GGTSTNGSDD252 pKa = 3.65YY253 pKa = 11.27NSTFTFSNGVIDD265 pKa = 5.01NGDD268 pKa = 3.38GTITVPGGVSSFTITASTTDD288 pKa = 3.44DD289 pKa = 3.38SSIEE293 pKa = 4.1STEE296 pKa = 3.95TLILNIGSKK305 pKa = 10.2SDD307 pKa = 3.3SGNILDD313 pKa = 4.5NDD315 pKa = 4.22SVALTASTSQTNLFCFGGSNGTASVVVSGGTAPYY349 pKa = 9.62SYY351 pKa = 10.44SWSPSGGTWATASGLTAGNYY371 pKa = 7.39TVTITDD377 pKa = 4.36DD378 pKa = 3.47NLATITKK385 pKa = 9.28NFTITEE391 pKa = 4.32PSAITASASQTNVSCNGGSNGTASVTPSGGAGSYY425 pKa = 9.0TYY427 pKa = 10.52SWSPSGGTGATASGLSAGSYY447 pKa = 8.99TVTIEE452 pKa = 4.44DD453 pKa = 4.54ANGCSATRR461 pKa = 11.84NFTITEE467 pKa = 4.26PSAIAASASQTNIACNGGATGSASVTPSGGAGSYY501 pKa = 9.0TYY503 pKa = 10.52SWSPSGGTGATASGLTAGNYY523 pKa = 7.39TVTITDD529 pKa = 4.04ANLCTATRR537 pKa = 11.84NFTITEE543 pKa = 4.3PSAITATTSQTNTSCNGSTDD563 pKa = 4.29GIASVIASGGTGSYY577 pKa = 10.04SYY579 pKa = 10.72SWSPSGGTAATATGLAAGNYY599 pKa = 6.88TCTITDD605 pKa = 4.26LNGCSIDD612 pKa = 3.67KK613 pKa = 10.63NFTIALSSSWNDD625 pKa = 3.15FDD627 pKa = 6.33CDD629 pKa = 3.75GDD631 pKa = 4.08GVTNGTEE638 pKa = 4.25VTDD641 pKa = 3.82GTNPDD646 pKa = 3.49NSCSFTASSITLNASSSWNTADD668 pKa = 4.57CDD670 pKa = 3.83GDD672 pKa = 3.79NVTNGQEE679 pKa = 4.31LLDD682 pKa = 3.99GTDD685 pKa = 3.54PNKK688 pKa = 10.31PDD690 pKa = 3.53TDD692 pKa = 3.91GDD694 pKa = 4.18GVTDD698 pKa = 4.12GKK700 pKa = 11.16EE701 pKa = 4.01KK702 pKa = 10.28TDD704 pKa = 3.48EE705 pKa = 4.09TDD707 pKa = 3.02ATDD710 pKa = 3.36PCKK713 pKa = 10.28FVFASQSVAPSSSWNTADD731 pKa = 4.57CDD733 pKa = 3.83GDD735 pKa = 3.79NVTNGQEE742 pKa = 4.27KK743 pKa = 10.89LDD745 pKa = 3.92GTDD748 pKa = 3.28SNKK751 pKa = 9.89PDD753 pKa = 3.57TDD755 pKa = 3.69GDD757 pKa = 4.18GVTDD761 pKa = 4.15GKK763 pKa = 11.02EE764 pKa = 3.86KK765 pKa = 10.41TDD767 pKa = 3.4ATDD770 pKa = 3.31ATDD773 pKa = 3.32PCKK776 pKa = 10.28FVFASQSVTPSSSWNTADD794 pKa = 5.11CDD796 pKa = 4.05GDD798 pKa = 4.11TITNGQEE805 pKa = 4.16KK806 pKa = 10.68LDD808 pKa = 3.97GTDD811 pKa = 3.21PTKK814 pKa = 10.67PDD816 pKa = 3.39TDD818 pKa = 3.89GDD820 pKa = 4.23GVTDD824 pKa = 4.64DD825 pKa = 5.05KK826 pKa = 11.61EE827 pKa = 4.31KK828 pKa = 10.65TDD830 pKa = 3.74ATDD833 pKa = 3.31ATDD836 pKa = 3.32PCKK839 pKa = 10.28FVFASQSVAPSSSWNTGDD857 pKa = 4.56CDD859 pKa = 4.26GDD861 pKa = 3.69NVTNGQEE868 pKa = 4.31KK869 pKa = 10.86LDD871 pKa = 3.9GTDD874 pKa = 3.66PNVADD879 pKa = 3.92TDD881 pKa = 4.2GDD883 pKa = 4.15GVTDD887 pKa = 4.39DD888 pKa = 4.71KK889 pKa = 11.71EE890 pKa = 4.39KK891 pKa = 10.69TDD893 pKa = 3.48VTDD896 pKa = 4.27ATDD899 pKa = 3.19ACQFILASQSVAPRR913 pKa = 11.84SSWNTADD920 pKa = 4.11CDD922 pKa = 3.74GDD924 pKa = 3.84NVSNGQEE931 pKa = 4.5KK932 pKa = 10.02IDD934 pKa = 3.74GTDD937 pKa = 3.35PLKK940 pKa = 10.77PDD942 pKa = 3.44TDD944 pKa = 4.09GDD946 pKa = 4.28GVTDD950 pKa = 3.83GTEE953 pKa = 4.41KK954 pKa = 10.18IDD956 pKa = 3.47GTNAKK961 pKa = 7.86NTCDD965 pKa = 3.67YY966 pKa = 10.94LAASQTLSPSALWNDD981 pKa = 3.76ADD983 pKa = 4.86CDD985 pKa = 3.67NDD987 pKa = 4.54GIRR990 pKa = 11.84NGDD993 pKa = 3.87DD994 pKa = 3.07NCVFTANANQTDD1006 pKa = 3.75NDD1008 pKa = 3.73NDD1010 pKa = 4.27GLGDD1014 pKa = 3.73TCDD1017 pKa = 5.63DD1018 pKa = 5.02DD1019 pKa = 6.69DD1020 pKa = 6.85DD1021 pKa = 5.98NDD1023 pKa = 5.22GILDD1027 pKa = 3.97TVDD1030 pKa = 3.44NCPLTPNSDD1039 pKa = 3.47QADD1042 pKa = 3.73RR1043 pKa = 11.84DD1044 pKa = 3.76HH1045 pKa = 7.53DD1046 pKa = 4.32GKK1048 pKa = 11.69GDD1050 pKa = 3.66VCDD1053 pKa = 4.54LGLNVSQAITPNGDD1067 pKa = 3.68GINDD1071 pKa = 2.8TWMIYY1076 pKa = 10.4NIEE1079 pKa = 4.17QYY1081 pKa = 10.21PNSTVRR1087 pKa = 11.84VFNRR1091 pKa = 11.84WGSEE1095 pKa = 2.89IFYY1098 pKa = 10.57AHH1100 pKa = 7.43NYY1102 pKa = 8.72RR1103 pKa = 11.84NDD1105 pKa = 3.27WDD1107 pKa = 3.59GHH1109 pKa = 4.92YY1110 pKa = 10.65KK1111 pKa = 10.53NKK1113 pKa = 9.95NQSLPEE1119 pKa = 3.89TSAYY1123 pKa = 10.27YY1124 pKa = 9.61YY1125 pKa = 10.72QIDD1128 pKa = 3.52IEE1130 pKa = 4.82GNGSVDD1136 pKa = 3.77LEE1138 pKa = 3.89GWIYY1142 pKa = 9.21LTKK1145 pKa = 10.84
Molecular weight: 118.34 kDa
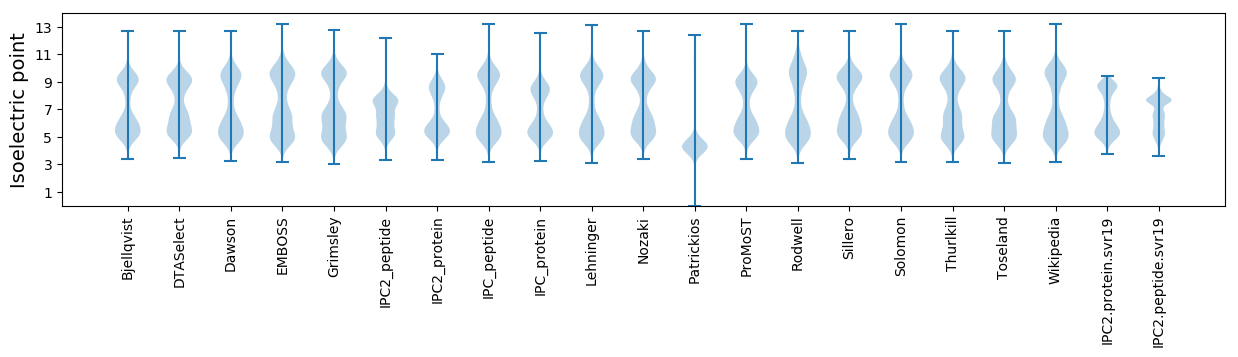
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A495SIM6|A0A495SIM6_9FLAO UDP-N-acetylglucosamine 2-epimerase (Hydrolysing) OS=Flavobacterium sp. 123 OX=2135627 GN=C8C88_1831 PE=4 SV=1
MM1 pKa = 7.84PSGKK5 pKa = 9.32KK6 pKa = 9.59RR7 pKa = 11.84KK8 pKa = 7.05RR9 pKa = 11.84HH10 pKa = 5.11KK11 pKa = 10.59VATHH15 pKa = 5.35KK16 pKa = 10.31RR17 pKa = 11.84KK18 pKa = 9.82KK19 pKa = 8.65RR20 pKa = 11.84ARR22 pKa = 11.84ANRR25 pKa = 11.84HH26 pKa = 4.91KK27 pKa = 10.59KK28 pKa = 9.82KK29 pKa = 10.51KK30 pKa = 10.04
MM1 pKa = 7.84PSGKK5 pKa = 9.32KK6 pKa = 9.59RR7 pKa = 11.84KK8 pKa = 7.05RR9 pKa = 11.84HH10 pKa = 5.11KK11 pKa = 10.59VATHH15 pKa = 5.35KK16 pKa = 10.31RR17 pKa = 11.84KK18 pKa = 9.82KK19 pKa = 8.65RR20 pKa = 11.84ARR22 pKa = 11.84ANRR25 pKa = 11.84HH26 pKa = 4.91KK27 pKa = 10.59KK28 pKa = 9.82KK29 pKa = 10.51KK30 pKa = 10.04
Molecular weight: 3.68 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
879023 |
30 |
4022 |
349.8 |
39.29 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.589 ± 0.057 | 0.797 ± 0.02 |
5.246 ± 0.036 | 6.304 ± 0.058 |
5.326 ± 0.045 | 6.43 ± 0.059 |
1.654 ± 0.025 | 8.435 ± 0.048 |
7.969 ± 0.072 | 9.134 ± 0.058 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.24 ± 0.028 | 6.265 ± 0.052 |
3.389 ± 0.028 | 3.329 ± 0.026 |
3.039 ± 0.035 | 6.642 ± 0.057 |
6.031 ± 0.111 | 6.271 ± 0.045 |
0.991 ± 0.018 | 3.92 ± 0.034 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
