
Lake Sarah-associated circular virus-7
Taxonomy: Viruses; unclassified viruses
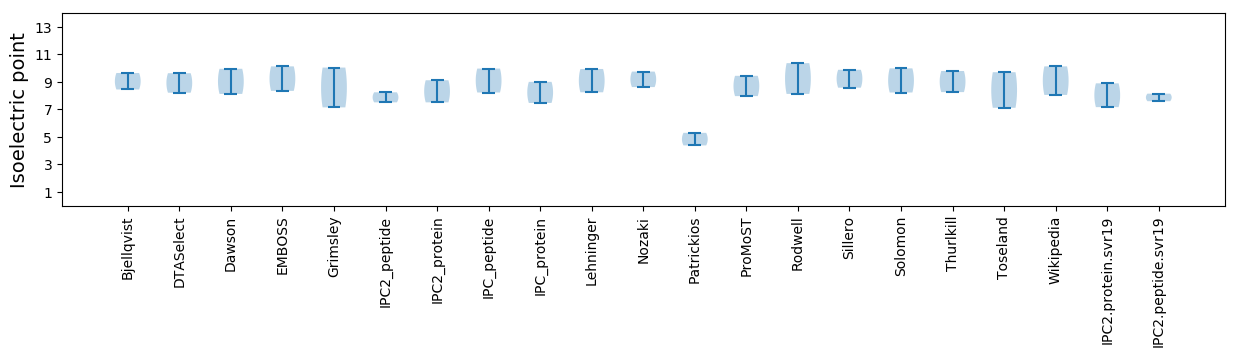
Average proteome isoelectric point is 8.03
Get precalculated fractions of proteins
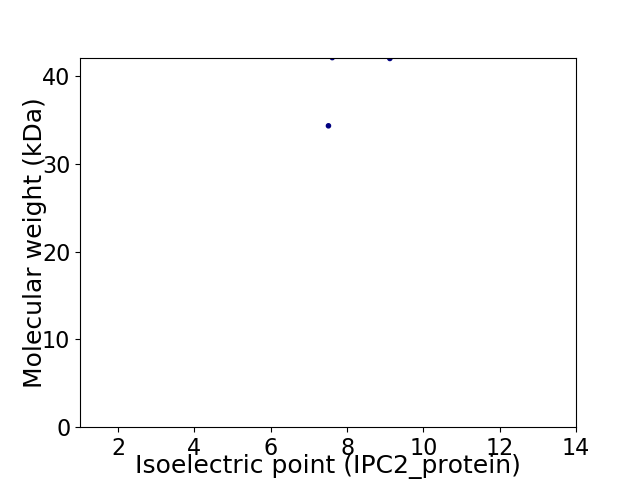
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A126G9J8|A0A126G9J8_9VIRU ATP-dependent helicase Rep OS=Lake Sarah-associated circular virus-7 OX=1685784 PE=3 SV=1
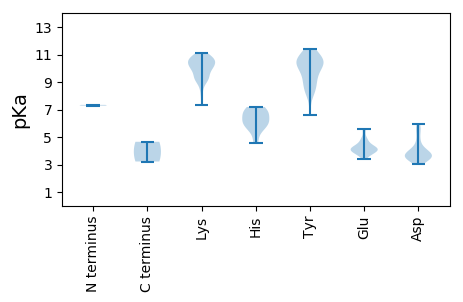
MM1 pKa = 7.28FCYY4 pKa = 10.02IKK6 pKa = 10.83NAISCLRR13 pKa = 11.84TVKK16 pKa = 10.47INVFSMPPSRR26 pKa = 11.84SRR28 pKa = 11.84AFIFTWNNPTADD40 pKa = 3.45TEE42 pKa = 4.25AALEE46 pKa = 4.11SLAGYY51 pKa = 10.25SYY53 pKa = 10.45LTFGRR58 pKa = 11.84EE59 pKa = 3.97TAPTTGTRR67 pKa = 11.84HH68 pKa = 4.55LQGYY72 pKa = 9.48IRR74 pKa = 11.84FTDD77 pKa = 3.97GKK79 pKa = 8.54SLRR82 pKa = 11.84SARR85 pKa = 11.84RR86 pKa = 11.84LLNGAHH92 pKa = 5.81VEE94 pKa = 4.11VARR97 pKa = 11.84TIRR100 pKa = 11.84QAIEE104 pKa = 3.97YY105 pKa = 8.48CHH107 pKa = 6.85KK108 pKa = 10.79EE109 pKa = 3.42GDD111 pKa = 3.73FVEE114 pKa = 5.5FGTRR118 pKa = 11.84PVDD121 pKa = 3.44DD122 pKa = 4.14AARR125 pKa = 11.84GDD127 pKa = 3.69MEE129 pKa = 4.09KK130 pKa = 10.76ARR132 pKa = 11.84WEE134 pKa = 4.21IAWTKK139 pKa = 11.1AKK141 pKa = 9.47TADD144 pKa = 4.21LEE146 pKa = 4.78EE147 pKa = 5.58IDD149 pKa = 3.39ADD151 pKa = 3.45IRR153 pKa = 11.84VRR155 pKa = 11.84CYY157 pKa = 10.64SALTRR162 pKa = 11.84IQKK165 pKa = 10.71DD166 pKa = 3.71YY167 pKa = 10.45MVPPLPLPAPCGLWIHH183 pKa = 6.2GLSGVGKK190 pKa = 8.13TFAVYY195 pKa = 9.37QAYY198 pKa = 9.24PDD200 pKa = 4.75LYY202 pKa = 11.09SKK204 pKa = 10.72NASKK208 pKa = 9.19WWDD211 pKa = 3.54GYY213 pKa = 10.29QNQDD217 pKa = 3.67HH218 pKa = 6.88ILFDD222 pKa = 5.38DD223 pKa = 3.9MDD225 pKa = 5.64PDD227 pKa = 3.56VGKK230 pKa = 9.39WAGRR234 pKa = 11.84FFKK237 pKa = 10.04IWADD241 pKa = 3.57EE242 pKa = 4.29RR243 pKa = 11.84PFIADD248 pKa = 3.06IKK250 pKa = 10.43GGSISIRR257 pKa = 11.84PKK259 pKa = 10.06IFIVTSQYY267 pKa = 11.0TIDD270 pKa = 3.52EE271 pKa = 4.61CFGEE275 pKa = 4.21IQTRR279 pKa = 11.84MALSRR284 pKa = 11.84RR285 pKa = 11.84FRR287 pKa = 11.84IIEE290 pKa = 4.15KK291 pKa = 10.42LSLDD295 pKa = 3.55HH296 pKa = 7.19EE297 pKa = 4.92INII300 pKa = 4.65
MM1 pKa = 7.28FCYY4 pKa = 10.02IKK6 pKa = 10.83NAISCLRR13 pKa = 11.84TVKK16 pKa = 10.47INVFSMPPSRR26 pKa = 11.84SRR28 pKa = 11.84AFIFTWNNPTADD40 pKa = 3.45TEE42 pKa = 4.25AALEE46 pKa = 4.11SLAGYY51 pKa = 10.25SYY53 pKa = 10.45LTFGRR58 pKa = 11.84EE59 pKa = 3.97TAPTTGTRR67 pKa = 11.84HH68 pKa = 4.55LQGYY72 pKa = 9.48IRR74 pKa = 11.84FTDD77 pKa = 3.97GKK79 pKa = 8.54SLRR82 pKa = 11.84SARR85 pKa = 11.84RR86 pKa = 11.84LLNGAHH92 pKa = 5.81VEE94 pKa = 4.11VARR97 pKa = 11.84TIRR100 pKa = 11.84QAIEE104 pKa = 3.97YY105 pKa = 8.48CHH107 pKa = 6.85KK108 pKa = 10.79EE109 pKa = 3.42GDD111 pKa = 3.73FVEE114 pKa = 5.5FGTRR118 pKa = 11.84PVDD121 pKa = 3.44DD122 pKa = 4.14AARR125 pKa = 11.84GDD127 pKa = 3.69MEE129 pKa = 4.09KK130 pKa = 10.76ARR132 pKa = 11.84WEE134 pKa = 4.21IAWTKK139 pKa = 11.1AKK141 pKa = 9.47TADD144 pKa = 4.21LEE146 pKa = 4.78EE147 pKa = 5.58IDD149 pKa = 3.39ADD151 pKa = 3.45IRR153 pKa = 11.84VRR155 pKa = 11.84CYY157 pKa = 10.64SALTRR162 pKa = 11.84IQKK165 pKa = 10.71DD166 pKa = 3.71YY167 pKa = 10.45MVPPLPLPAPCGLWIHH183 pKa = 6.2GLSGVGKK190 pKa = 8.13TFAVYY195 pKa = 9.37QAYY198 pKa = 9.24PDD200 pKa = 4.75LYY202 pKa = 11.09SKK204 pKa = 10.72NASKK208 pKa = 9.19WWDD211 pKa = 3.54GYY213 pKa = 10.29QNQDD217 pKa = 3.67HH218 pKa = 6.88ILFDD222 pKa = 5.38DD223 pKa = 3.9MDD225 pKa = 5.64PDD227 pKa = 3.56VGKK230 pKa = 9.39WAGRR234 pKa = 11.84FFKK237 pKa = 10.04IWADD241 pKa = 3.57EE242 pKa = 4.29RR243 pKa = 11.84PFIADD248 pKa = 3.06IKK250 pKa = 10.43GGSISIRR257 pKa = 11.84PKK259 pKa = 10.06IFIVTSQYY267 pKa = 11.0TIDD270 pKa = 3.52EE271 pKa = 4.61CFGEE275 pKa = 4.21IQTRR279 pKa = 11.84MALSRR284 pKa = 11.84RR285 pKa = 11.84FRR287 pKa = 11.84IIEE290 pKa = 4.15KK291 pKa = 10.42LSLDD295 pKa = 3.55HH296 pKa = 7.19EE297 pKa = 4.92INII300 pKa = 4.65
Molecular weight: 34.32 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A126G9J8|A0A126G9J8_9VIRU ATP-dependent helicase Rep OS=Lake Sarah-associated circular virus-7 OX=1685784 PE=3 SV=1
MM1 pKa = 7.34GRR3 pKa = 11.84RR4 pKa = 11.84FRR6 pKa = 11.84KK7 pKa = 9.01RR8 pKa = 11.84GKK10 pKa = 10.17VIQTTPQGQRR20 pKa = 11.84TIPYY24 pKa = 8.2NASASRR30 pKa = 11.84AAFKK34 pKa = 10.97VADD37 pKa = 4.41AAGKK41 pKa = 9.14WLGKK45 pKa = 9.27KK46 pKa = 9.9AKK48 pKa = 9.98AYY50 pKa = 8.26WNSPTPKK57 pKa = 9.93VYY59 pKa = 10.3KK60 pKa = 10.28KK61 pKa = 10.16KK62 pKa = 10.67SKK64 pKa = 10.18PKK66 pKa = 9.4EE67 pKa = 3.79EE68 pKa = 3.97AKK70 pKa = 10.44EE71 pKa = 3.7LHH73 pKa = 6.12TSVNGGGNVINTIVNIGKK91 pKa = 9.52SNKK94 pKa = 9.21KK95 pKa = 7.3QAKK98 pKa = 9.24LIKK101 pKa = 10.24KK102 pKa = 9.45LGAYY106 pKa = 6.59TTSQYY111 pKa = 11.18LAAFTHH117 pKa = 6.15VNTATEE123 pKa = 3.93ASTQEE128 pKa = 4.13VNDD131 pKa = 3.3VAVYY135 pKa = 10.37LKK137 pKa = 10.65GQSLWDD143 pKa = 3.42IVQNAYY149 pKa = 8.36QQNAAWQALAPNLTTAGAQARR170 pKa = 11.84IYY172 pKa = 10.56VSYY175 pKa = 10.48IKK177 pKa = 10.04TEE179 pKa = 3.79IEE181 pKa = 4.12FVNLSPAICFVDD193 pKa = 3.89IYY195 pKa = 10.84LFRR198 pKa = 11.84AKK200 pKa = 10.13VDD202 pKa = 3.35QLAFSDD208 pKa = 3.83VTSAYY213 pKa = 10.45NLPSNTWATSIDD225 pKa = 3.68QEE227 pKa = 4.35KK228 pKa = 10.91GLGSVSQTFPGMEE241 pKa = 3.71PRR243 pKa = 11.84GALFKK248 pKa = 10.92KK249 pKa = 9.84HH250 pKa = 5.87FKK252 pKa = 9.97EE253 pKa = 4.48VYY255 pKa = 10.02KK256 pKa = 10.6LRR258 pKa = 11.84CCMNGGEE265 pKa = 4.23VRR267 pKa = 11.84RR268 pKa = 11.84LSIYY272 pKa = 8.85MHH274 pKa = 5.48MNKK277 pKa = 8.95WVEE280 pKa = 3.94SAKK283 pKa = 10.53IILNKK288 pKa = 10.53GNLNGLTHH296 pKa = 6.91HH297 pKa = 6.69LMFVEE302 pKa = 4.82RR303 pKa = 11.84GSPCGNLNDD312 pKa = 4.97PYY314 pKa = 11.4VADD317 pKa = 3.83TQVYY321 pKa = 9.84LSPIKK326 pKa = 10.7INGTVNTTFGVGCLVRR342 pKa = 11.84PSNNAYY348 pKa = 9.26TSQTGLTTSYY358 pKa = 10.64AANVYY363 pKa = 8.85TLVDD367 pKa = 3.98DD368 pKa = 5.92DD369 pKa = 5.67GGTKK373 pKa = 10.56NIGVVDD379 pKa = 3.67SFGG382 pKa = 3.22
MM1 pKa = 7.34GRR3 pKa = 11.84RR4 pKa = 11.84FRR6 pKa = 11.84KK7 pKa = 9.01RR8 pKa = 11.84GKK10 pKa = 10.17VIQTTPQGQRR20 pKa = 11.84TIPYY24 pKa = 8.2NASASRR30 pKa = 11.84AAFKK34 pKa = 10.97VADD37 pKa = 4.41AAGKK41 pKa = 9.14WLGKK45 pKa = 9.27KK46 pKa = 9.9AKK48 pKa = 9.98AYY50 pKa = 8.26WNSPTPKK57 pKa = 9.93VYY59 pKa = 10.3KK60 pKa = 10.28KK61 pKa = 10.16KK62 pKa = 10.67SKK64 pKa = 10.18PKK66 pKa = 9.4EE67 pKa = 3.79EE68 pKa = 3.97AKK70 pKa = 10.44EE71 pKa = 3.7LHH73 pKa = 6.12TSVNGGGNVINTIVNIGKK91 pKa = 9.52SNKK94 pKa = 9.21KK95 pKa = 7.3QAKK98 pKa = 9.24LIKK101 pKa = 10.24KK102 pKa = 9.45LGAYY106 pKa = 6.59TTSQYY111 pKa = 11.18LAAFTHH117 pKa = 6.15VNTATEE123 pKa = 3.93ASTQEE128 pKa = 4.13VNDD131 pKa = 3.3VAVYY135 pKa = 10.37LKK137 pKa = 10.65GQSLWDD143 pKa = 3.42IVQNAYY149 pKa = 8.36QQNAAWQALAPNLTTAGAQARR170 pKa = 11.84IYY172 pKa = 10.56VSYY175 pKa = 10.48IKK177 pKa = 10.04TEE179 pKa = 3.79IEE181 pKa = 4.12FVNLSPAICFVDD193 pKa = 3.89IYY195 pKa = 10.84LFRR198 pKa = 11.84AKK200 pKa = 10.13VDD202 pKa = 3.35QLAFSDD208 pKa = 3.83VTSAYY213 pKa = 10.45NLPSNTWATSIDD225 pKa = 3.68QEE227 pKa = 4.35KK228 pKa = 10.91GLGSVSQTFPGMEE241 pKa = 3.71PRR243 pKa = 11.84GALFKK248 pKa = 10.92KK249 pKa = 9.84HH250 pKa = 5.87FKK252 pKa = 9.97EE253 pKa = 4.48VYY255 pKa = 10.02KK256 pKa = 10.6LRR258 pKa = 11.84CCMNGGEE265 pKa = 4.23VRR267 pKa = 11.84RR268 pKa = 11.84LSIYY272 pKa = 8.85MHH274 pKa = 5.48MNKK277 pKa = 8.95WVEE280 pKa = 3.94SAKK283 pKa = 10.53IILNKK288 pKa = 10.53GNLNGLTHH296 pKa = 6.91HH297 pKa = 6.69LMFVEE302 pKa = 4.82RR303 pKa = 11.84GSPCGNLNDD312 pKa = 4.97PYY314 pKa = 11.4VADD317 pKa = 3.83TQVYY321 pKa = 9.84LSPIKK326 pKa = 10.7INGTVNTTFGVGCLVRR342 pKa = 11.84PSNNAYY348 pKa = 9.26TSQTGLTTSYY358 pKa = 10.64AANVYY363 pKa = 8.85TLVDD367 pKa = 3.98DD368 pKa = 5.92DD369 pKa = 5.67GGTKK373 pKa = 10.56NIGVVDD379 pKa = 3.67SFGG382 pKa = 3.22
Molecular weight: 41.98 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
682 |
300 |
382 |
341.0 |
38.15 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.238 ± 0.152 | 1.613 ± 0.248 |
4.985 ± 1.29 | 4.252 ± 0.692 |
4.252 ± 0.692 | 7.185 ± 0.545 |
1.76 ± 0.154 | 6.598 ± 1.325 |
7.185 ± 1.186 | 6.598 ± 0.17 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.76 ± 0.154 | 5.279 ± 1.673 |
4.106 ± 0.359 | 3.666 ± 0.64 |
5.572 ± 1.555 | 6.158 ± 0.315 |
7.185 ± 0.545 | 6.158 ± 1.382 |
2.053 ± 0.393 | 4.399 ± 0.255 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
