
Apis cerana cerana (Oriental honeybee)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Protostomia; Ecdysozoa; Panarthropoda; Arthropoda; Mandibulata; Pancrustacea; Hexapoda; Insecta; Dicondylia; Pterygota; Neoptera; Endopterygota; Hymenoptera; Apocrita; Aculeata; Apoidea; Apidae; Apinae; Apini;
Average proteome isoelectric point is 6.83
Get precalculated fractions of proteins
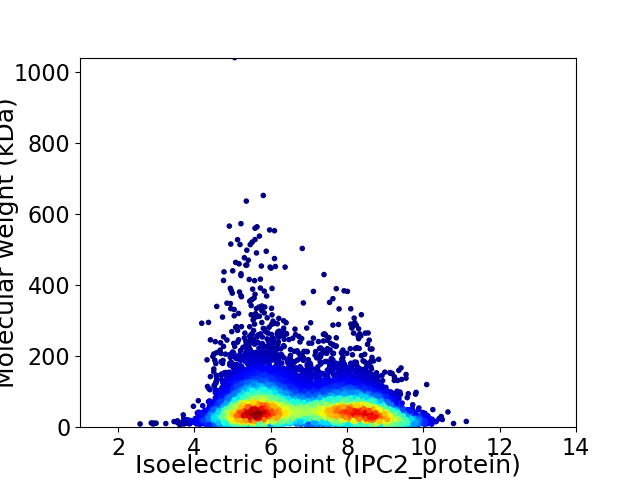
Virtual 2D-PAGE plot for 9931 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2A3ECG4|A0A2A3ECG4_APICC Iporin OS=Apis cerana cerana OX=94128 GN=APICC_06640 PE=4 SV=1
MM1 pKa = 7.11VVLSNFLAPQEE12 pKa = 4.56GIRR15 pKa = 11.84HH16 pKa = 5.6EE17 pKa = 4.32EE18 pKa = 3.96QNTTVYY24 pKa = 10.76INDD27 pKa = 3.7RR28 pKa = 11.84EE29 pKa = 4.38VGKK32 pKa = 8.77GTLYY36 pKa = 9.3ITEE39 pKa = 4.71SLLSWVNYY47 pKa = 7.76DD48 pKa = 3.57TQQGFSLEE56 pKa = 4.16YY57 pKa = 10.02PHH59 pKa = 7.27ISLHH63 pKa = 6.32AISRR67 pKa = 11.84DD68 pKa = 3.7EE69 pKa = 3.97QVHH72 pKa = 5.94PRR74 pKa = 11.84QCLYY78 pKa = 10.85IMVDD82 pKa = 3.51AKK84 pKa = 11.31VDD86 pKa = 3.89LPDD89 pKa = 3.65VSLSPASDD97 pKa = 3.15SGSEE101 pKa = 4.23NEE103 pKa = 5.53FEE105 pKa = 5.55DD106 pKa = 4.25ADD108 pKa = 4.15TPITEE113 pKa = 4.27MRR115 pKa = 11.84FAPDD119 pKa = 3.02NTNNLEE125 pKa = 4.54AMFQAMNQCQALHH138 pKa = 7.44PDD140 pKa = 3.88PQDD143 pKa = 3.37SFSDD147 pKa = 3.74AEE149 pKa = 3.83EE150 pKa = 5.32DD151 pKa = 3.31IYY153 pKa = 11.51EE154 pKa = 4.54DD155 pKa = 4.26AEE157 pKa = 4.14EE158 pKa = 5.55DD159 pKa = 3.94DD160 pKa = 4.26FEE162 pKa = 6.75HH163 pKa = 7.1YY164 pKa = 10.65DD165 pKa = 3.6VGAGDD170 pKa = 4.46APYY173 pKa = 10.4ILPTGFLFSFYY184 pKa = 10.87NILEE188 pKa = 4.37QIGTNHH194 pKa = 6.66NGTEE198 pKa = 4.34ADD200 pKa = 3.46DD201 pKa = 4.6AMDD204 pKa = 3.71IEE206 pKa = 4.99AGQFEE211 pKa = 4.76DD212 pKa = 5.49AEE214 pKa = 4.39EE215 pKa = 4.16DD216 pKa = 3.59LL217 pKa = 5.34
MM1 pKa = 7.11VVLSNFLAPQEE12 pKa = 4.56GIRR15 pKa = 11.84HH16 pKa = 5.6EE17 pKa = 4.32EE18 pKa = 3.96QNTTVYY24 pKa = 10.76INDD27 pKa = 3.7RR28 pKa = 11.84EE29 pKa = 4.38VGKK32 pKa = 8.77GTLYY36 pKa = 9.3ITEE39 pKa = 4.71SLLSWVNYY47 pKa = 7.76DD48 pKa = 3.57TQQGFSLEE56 pKa = 4.16YY57 pKa = 10.02PHH59 pKa = 7.27ISLHH63 pKa = 6.32AISRR67 pKa = 11.84DD68 pKa = 3.7EE69 pKa = 3.97QVHH72 pKa = 5.94PRR74 pKa = 11.84QCLYY78 pKa = 10.85IMVDD82 pKa = 3.51AKK84 pKa = 11.31VDD86 pKa = 3.89LPDD89 pKa = 3.65VSLSPASDD97 pKa = 3.15SGSEE101 pKa = 4.23NEE103 pKa = 5.53FEE105 pKa = 5.55DD106 pKa = 4.25ADD108 pKa = 4.15TPITEE113 pKa = 4.27MRR115 pKa = 11.84FAPDD119 pKa = 3.02NTNNLEE125 pKa = 4.54AMFQAMNQCQALHH138 pKa = 7.44PDD140 pKa = 3.88PQDD143 pKa = 3.37SFSDD147 pKa = 3.74AEE149 pKa = 3.83EE150 pKa = 5.32DD151 pKa = 3.31IYY153 pKa = 11.51EE154 pKa = 4.54DD155 pKa = 4.26AEE157 pKa = 4.14EE158 pKa = 5.55DD159 pKa = 3.94DD160 pKa = 4.26FEE162 pKa = 6.75HH163 pKa = 7.1YY164 pKa = 10.65DD165 pKa = 3.6VGAGDD170 pKa = 4.46APYY173 pKa = 10.4ILPTGFLFSFYY184 pKa = 10.87NILEE188 pKa = 4.37QIGTNHH194 pKa = 6.66NGTEE198 pKa = 4.34ADD200 pKa = 3.46DD201 pKa = 4.6AMDD204 pKa = 3.71IEE206 pKa = 4.99AGQFEE211 pKa = 4.76DD212 pKa = 5.49AEE214 pKa = 4.39EE215 pKa = 4.16DD216 pKa = 3.59LL217 pKa = 5.34
Molecular weight: 24.45 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2A3EPC0|A0A2A3EPC0_APICC Abasic site processing protein HMCES OS=Apis cerana cerana OX=94128 GN=APICC_01750 PE=3 SV=1
MM1 pKa = 7.47SPLNRR6 pKa = 11.84GPLLGPRR13 pKa = 11.84VPPTGVRR20 pKa = 11.84PPPPPRR26 pKa = 11.84FGGRR30 pKa = 11.84LPPPGLPPRR39 pKa = 11.84MPPPPMNPVFGPMRR53 pKa = 11.84QRR55 pKa = 11.84LPPPPRR61 pKa = 11.84PGGVNRR67 pKa = 11.84PSGPMPLFGPRR78 pKa = 11.84VRR80 pKa = 11.84AMAPMVPPMGLRR92 pKa = 11.84GVGPRR97 pKa = 11.84GPLMRR102 pKa = 11.84PWHH105 pKa = 6.23RR106 pKa = 11.84RR107 pKa = 11.84ALPPQILSHH116 pKa = 5.24MRR118 pKa = 11.84PRR120 pKa = 11.84FSIRR124 pKa = 11.84NGNVKK129 pKa = 9.9GKK131 pKa = 10.7AMNNAKK137 pKa = 10.02KK138 pKa = 10.5VSKK141 pKa = 10.99LEE143 pKa = 4.0NNYY146 pKa = 9.76VKK148 pKa = 10.89
MM1 pKa = 7.47SPLNRR6 pKa = 11.84GPLLGPRR13 pKa = 11.84VPPTGVRR20 pKa = 11.84PPPPPRR26 pKa = 11.84FGGRR30 pKa = 11.84LPPPGLPPRR39 pKa = 11.84MPPPPMNPVFGPMRR53 pKa = 11.84QRR55 pKa = 11.84LPPPPRR61 pKa = 11.84PGGVNRR67 pKa = 11.84PSGPMPLFGPRR78 pKa = 11.84VRR80 pKa = 11.84AMAPMVPPMGLRR92 pKa = 11.84GVGPRR97 pKa = 11.84GPLMRR102 pKa = 11.84PWHH105 pKa = 6.23RR106 pKa = 11.84RR107 pKa = 11.84ALPPQILSHH116 pKa = 5.24MRR118 pKa = 11.84PRR120 pKa = 11.84FSIRR124 pKa = 11.84NGNVKK129 pKa = 9.9GKK131 pKa = 10.7AMNNAKK137 pKa = 10.02KK138 pKa = 10.5VSKK141 pKa = 10.99LEE143 pKa = 4.0NNYY146 pKa = 9.76VKK148 pKa = 10.89
Molecular weight: 16.2 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5621110 |
31 |
9157 |
566.0 |
64.16 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.481 ± 0.023 | 1.965 ± 0.019 |
5.285 ± 0.015 | 7.072 ± 0.035 |
3.66 ± 0.019 | 5.128 ± 0.03 |
2.425 ± 0.012 | 6.788 ± 0.028 |
6.999 ± 0.03 | 9.108 ± 0.03 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.272 ± 0.011 | 5.777 ± 0.024 |
4.693 ± 0.026 | 4.344 ± 0.025 |
5.19 ± 0.021 | 7.998 ± 0.029 |
5.892 ± 0.021 | 5.572 ± 0.016 |
1.078 ± 0.009 | 3.273 ± 0.015 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
