
Thiocapsa marina 5811
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Chromatiales; Chromatiaceae; Thiocapsa; Thiocapsa marina
Average proteome isoelectric point is 6.3
Get precalculated fractions of proteins
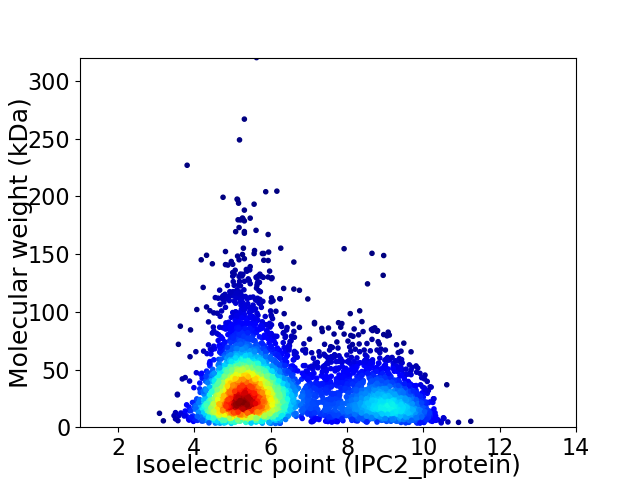
Virtual 2D-PAGE plot for 4915 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|F9UDC1|F9UDC1_9GAMM Sigma54 specific transcriptional regulator Fis family OS=Thiocapsa marina 5811 OX=768671 GN=ThimaDRAFT_2924 PE=4 SV=1
MM1 pKa = 7.07YY2 pKa = 9.98RR3 pKa = 11.84IKK5 pKa = 10.84HH6 pKa = 5.8PADD9 pKa = 3.96PSLVLAALTSGLLVASAAPSWAYY32 pKa = 10.7DD33 pKa = 3.32VTDD36 pKa = 3.85NFSIGAVLGAAGQCQEE52 pKa = 4.25GLGGLVDD59 pKa = 3.77EE60 pKa = 5.67AGQSLEE66 pKa = 4.17GDD68 pKa = 3.89TCRR71 pKa = 11.84GGMPFQLEE79 pKa = 4.03MSFRR83 pKa = 11.84PSEE86 pKa = 3.95RR87 pKa = 11.84DD88 pKa = 2.91EE89 pKa = 5.31LYY91 pKa = 11.07ALLGFAVDD99 pKa = 3.55NGLNAVSPFVLAPWAVDD116 pKa = 5.15LEE118 pKa = 5.72DD119 pKa = 5.52DD120 pKa = 4.26LQDD123 pKa = 3.14INGRR127 pKa = 11.84GRR129 pKa = 11.84DD130 pKa = 3.68YY131 pKa = 11.45LLQAWYY137 pKa = 10.01KK138 pKa = 8.84HH139 pKa = 4.88TFTLGEE145 pKa = 4.4DD146 pKa = 3.39ATLGATFGIIDD157 pKa = 3.57STGYY161 pKa = 10.79LDD163 pKa = 3.76EE164 pKa = 4.36NAYY167 pKa = 10.82ANDD170 pKa = 3.58EE171 pKa = 4.16FTQFMNEE178 pKa = 3.6AFVNSGGFNLPSYY191 pKa = 10.55DD192 pKa = 3.25AGAALEE198 pKa = 4.25LALGDD203 pKa = 3.89WEE205 pKa = 4.2ITAAALNIGEE215 pKa = 4.56NDD217 pKa = 3.99DD218 pKa = 3.82GNNYY222 pKa = 9.33NFWGAQLGYY231 pKa = 10.7RR232 pKa = 11.84LDD234 pKa = 3.7TALGEE239 pKa = 4.31GNYY242 pKa = 9.92RR243 pKa = 11.84AVITGTSSAFFKK255 pKa = 10.74PGAPLGEE262 pKa = 4.22EE263 pKa = 4.36PEE265 pKa = 4.35GTEE268 pKa = 5.07DD269 pKa = 4.13GSPLAAEE276 pKa = 4.48DD277 pKa = 4.25TPEE280 pKa = 4.05KK281 pKa = 9.48TDD283 pKa = 3.21LMGYY287 pKa = 9.96GLSFDD292 pKa = 3.54QALGDD297 pKa = 3.58RR298 pKa = 11.84LGAFLRR304 pKa = 11.84MSWTDD309 pKa = 3.28HH310 pKa = 6.52EE311 pKa = 6.2DD312 pKa = 3.16ILDD315 pKa = 3.64YY316 pKa = 11.01RR317 pKa = 11.84GLYY320 pKa = 9.2TGGLNIGGGLWGRR333 pKa = 11.84EE334 pKa = 3.64ADD336 pKa = 3.77NVGIAYY342 pKa = 9.61GYY344 pKa = 10.86LDD346 pKa = 3.95GANTGIANTNVAEE359 pKa = 4.09IYY361 pKa = 10.75YY362 pKa = 10.2RR363 pKa = 11.84FAANDD368 pKa = 3.74FLALTADD375 pKa = 3.69VQYY378 pKa = 10.55MADD381 pKa = 3.97AYY383 pKa = 11.0DD384 pKa = 3.53SGEE387 pKa = 4.89DD388 pKa = 3.25IEE390 pKa = 5.27GWIFGLRR397 pKa = 11.84LVAGLL402 pKa = 3.94
MM1 pKa = 7.07YY2 pKa = 9.98RR3 pKa = 11.84IKK5 pKa = 10.84HH6 pKa = 5.8PADD9 pKa = 3.96PSLVLAALTSGLLVASAAPSWAYY32 pKa = 10.7DD33 pKa = 3.32VTDD36 pKa = 3.85NFSIGAVLGAAGQCQEE52 pKa = 4.25GLGGLVDD59 pKa = 3.77EE60 pKa = 5.67AGQSLEE66 pKa = 4.17GDD68 pKa = 3.89TCRR71 pKa = 11.84GGMPFQLEE79 pKa = 4.03MSFRR83 pKa = 11.84PSEE86 pKa = 3.95RR87 pKa = 11.84DD88 pKa = 2.91EE89 pKa = 5.31LYY91 pKa = 11.07ALLGFAVDD99 pKa = 3.55NGLNAVSPFVLAPWAVDD116 pKa = 5.15LEE118 pKa = 5.72DD119 pKa = 5.52DD120 pKa = 4.26LQDD123 pKa = 3.14INGRR127 pKa = 11.84GRR129 pKa = 11.84DD130 pKa = 3.68YY131 pKa = 11.45LLQAWYY137 pKa = 10.01KK138 pKa = 8.84HH139 pKa = 4.88TFTLGEE145 pKa = 4.4DD146 pKa = 3.39ATLGATFGIIDD157 pKa = 3.57STGYY161 pKa = 10.79LDD163 pKa = 3.76EE164 pKa = 4.36NAYY167 pKa = 10.82ANDD170 pKa = 3.58EE171 pKa = 4.16FTQFMNEE178 pKa = 3.6AFVNSGGFNLPSYY191 pKa = 10.55DD192 pKa = 3.25AGAALEE198 pKa = 4.25LALGDD203 pKa = 3.89WEE205 pKa = 4.2ITAAALNIGEE215 pKa = 4.56NDD217 pKa = 3.99DD218 pKa = 3.82GNNYY222 pKa = 9.33NFWGAQLGYY231 pKa = 10.7RR232 pKa = 11.84LDD234 pKa = 3.7TALGEE239 pKa = 4.31GNYY242 pKa = 9.92RR243 pKa = 11.84AVITGTSSAFFKK255 pKa = 10.74PGAPLGEE262 pKa = 4.22EE263 pKa = 4.36PEE265 pKa = 4.35GTEE268 pKa = 5.07DD269 pKa = 4.13GSPLAAEE276 pKa = 4.48DD277 pKa = 4.25TPEE280 pKa = 4.05KK281 pKa = 9.48TDD283 pKa = 3.21LMGYY287 pKa = 9.96GLSFDD292 pKa = 3.54QALGDD297 pKa = 3.58RR298 pKa = 11.84LGAFLRR304 pKa = 11.84MSWTDD309 pKa = 3.28HH310 pKa = 6.52EE311 pKa = 6.2DD312 pKa = 3.16ILDD315 pKa = 3.64YY316 pKa = 11.01RR317 pKa = 11.84GLYY320 pKa = 9.2TGGLNIGGGLWGRR333 pKa = 11.84EE334 pKa = 3.64ADD336 pKa = 3.77NVGIAYY342 pKa = 9.61GYY344 pKa = 10.86LDD346 pKa = 3.95GANTGIANTNVAEE359 pKa = 4.09IYY361 pKa = 10.75YY362 pKa = 10.2RR363 pKa = 11.84FAANDD368 pKa = 3.74FLALTADD375 pKa = 3.69VQYY378 pKa = 10.55MADD381 pKa = 3.97AYY383 pKa = 11.0DD384 pKa = 3.53SGEE387 pKa = 4.89DD388 pKa = 3.25IEE390 pKa = 5.27GWIFGLRR397 pKa = 11.84LVAGLL402 pKa = 3.94
Molecular weight: 43.01 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|F9UCF1|F9UCF1_9GAMM ATP synthase subunit alpha OS=Thiocapsa marina 5811 OX=768671 GN=atpA PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.56RR3 pKa = 11.84TFQPSRR9 pKa = 11.84LKK11 pKa = 10.48RR12 pKa = 11.84ARR14 pKa = 11.84THH16 pKa = 5.93GFRR19 pKa = 11.84ARR21 pKa = 11.84SATKK25 pKa = 10.29NGRR28 pKa = 11.84KK29 pKa = 9.22VLNARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.49GRR39 pKa = 11.84VRR41 pKa = 11.84LATT44 pKa = 3.61
MM1 pKa = 7.45KK2 pKa = 9.56RR3 pKa = 11.84TFQPSRR9 pKa = 11.84LKK11 pKa = 10.48RR12 pKa = 11.84ARR14 pKa = 11.84THH16 pKa = 5.93GFRR19 pKa = 11.84ARR21 pKa = 11.84SATKK25 pKa = 10.29NGRR28 pKa = 11.84KK29 pKa = 9.22VLNARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.49GRR39 pKa = 11.84VRR41 pKa = 11.84LATT44 pKa = 3.61
Molecular weight: 5.11 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1589608 |
30 |
2892 |
323.4 |
35.44 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.571 ± 0.046 | 1.01 ± 0.013 |
6.032 ± 0.029 | 6.189 ± 0.031 |
3.472 ± 0.021 | 8.21 ± 0.031 |
2.169 ± 0.017 | 4.887 ± 0.021 |
2.533 ± 0.023 | 11.177 ± 0.048 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.193 ± 0.016 | 2.285 ± 0.019 |
5.567 ± 0.028 | 3.274 ± 0.022 |
8.147 ± 0.041 | 5.272 ± 0.023 |
5.053 ± 0.024 | 7.182 ± 0.032 |
1.447 ± 0.015 | 2.33 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
