
Simian T-lymphotropic virus 2
Taxonomy: Viruses; Riboviria; Pararnavirae; Artverviricota; Revtraviricetes; Ortervirales; Retroviridae; Orthoretrovirinae; Deltaretrovirus; Primate T-lymphotropic virus 2
Average proteome isoelectric point is 7.94
Get precalculated fractions of proteins
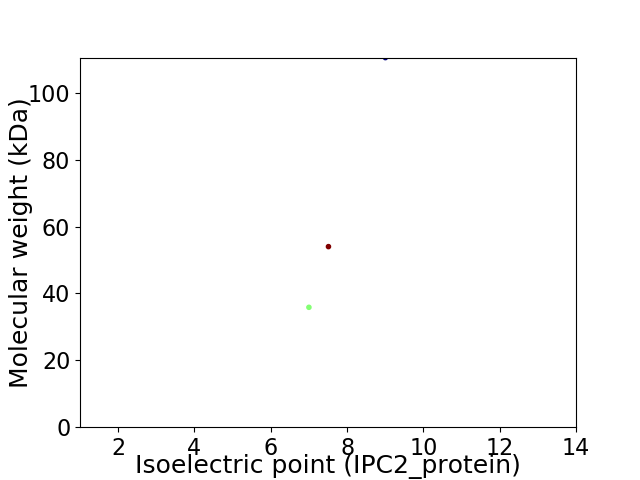
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
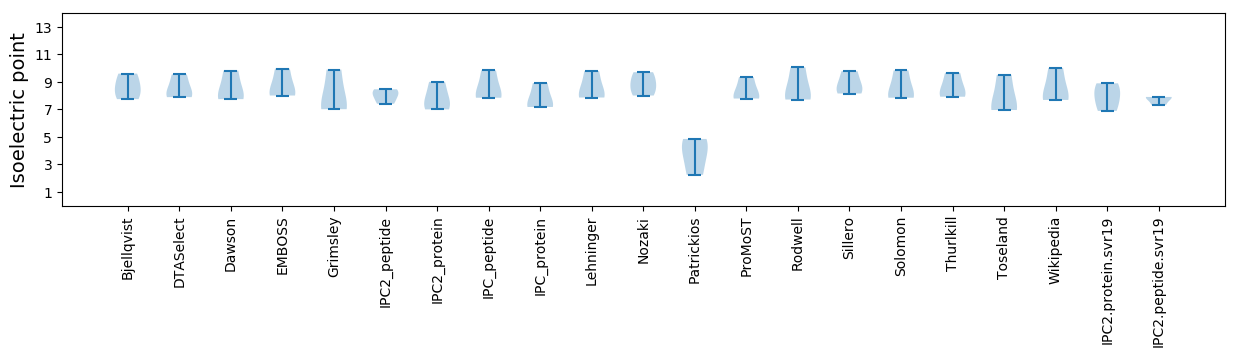
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|O09242|O09242_9DELA Pol protein (Fragment) OS=Simian T-lymphotropic virus 2 OX=33748 GN=pol PE=4 SV=1
MM1 pKa = 6.92GQTYY5 pKa = 10.25GLSPSPIPKK14 pKa = 10.02APRR17 pKa = 11.84GLSTHH22 pKa = 5.79HH23 pKa = 6.88WLNFLQASYY32 pKa = 10.71RR33 pKa = 11.84LQPGPSDD40 pKa = 4.38FDD42 pKa = 3.75FQQLRR47 pKa = 11.84RR48 pKa = 11.84FLKK51 pKa = 10.7LALKK55 pKa = 9.26TPIWLNPIDD64 pKa = 4.21YY65 pKa = 10.62SLLASLIPKK74 pKa = 8.85GYY76 pKa = 9.32PGRR79 pKa = 11.84TSEE82 pKa = 4.86IINVLIRR89 pKa = 11.84NQASPTPPPAPSLPEE104 pKa = 3.92PANPPPLQQPSAPPEE119 pKa = 3.9PHH121 pKa = 6.45TPPPYY126 pKa = 9.46IEE128 pKa = 5.11PPATHH133 pKa = 7.28CLPILHH139 pKa = 6.61PHH141 pKa = 6.84GAPSAHH147 pKa = 6.92RR148 pKa = 11.84PWQMKK153 pKa = 9.77DD154 pKa = 2.99LQAIKK159 pKa = 10.78QEE161 pKa = 4.53VNTSAPGSPQFMQTVRR177 pKa = 11.84LAIQQFDD184 pKa = 3.85PTAKK188 pKa = 10.47DD189 pKa = 3.57LQDD192 pKa = 3.86LLQYY196 pKa = 10.43LCSSLVVSLHH206 pKa = 5.5HH207 pKa = 5.89QQLHH211 pKa = 4.61TLITEE216 pKa = 4.28AEE218 pKa = 4.24TRR220 pKa = 11.84GMTGYY225 pKa = 11.08NPMAGPLRR233 pKa = 11.84MQANNPAQEE242 pKa = 4.26GLRR245 pKa = 11.84RR246 pKa = 11.84EE247 pKa = 4.29YY248 pKa = 11.06QNLWLAAFSALPGNTRR264 pKa = 11.84DD265 pKa = 4.02PSWAAILQGLEE276 pKa = 3.97EE277 pKa = 5.02PYY279 pKa = 10.54CALLEE284 pKa = 4.22RR285 pKa = 11.84LNVALDD291 pKa = 3.32NGLPEE296 pKa = 4.52GTPKK300 pKa = 10.76EE301 pKa = 4.23PILRR305 pKa = 11.84SLAYY309 pKa = 10.74SNANKK314 pKa = 10.2DD315 pKa = 3.9CQKK318 pKa = 10.69LAGSGPYY325 pKa = 9.91
MM1 pKa = 6.92GQTYY5 pKa = 10.25GLSPSPIPKK14 pKa = 10.02APRR17 pKa = 11.84GLSTHH22 pKa = 5.79HH23 pKa = 6.88WLNFLQASYY32 pKa = 10.71RR33 pKa = 11.84LQPGPSDD40 pKa = 4.38FDD42 pKa = 3.75FQQLRR47 pKa = 11.84RR48 pKa = 11.84FLKK51 pKa = 10.7LALKK55 pKa = 9.26TPIWLNPIDD64 pKa = 4.21YY65 pKa = 10.62SLLASLIPKK74 pKa = 8.85GYY76 pKa = 9.32PGRR79 pKa = 11.84TSEE82 pKa = 4.86IINVLIRR89 pKa = 11.84NQASPTPPPAPSLPEE104 pKa = 3.92PANPPPLQQPSAPPEE119 pKa = 3.9PHH121 pKa = 6.45TPPPYY126 pKa = 9.46IEE128 pKa = 5.11PPATHH133 pKa = 7.28CLPILHH139 pKa = 6.61PHH141 pKa = 6.84GAPSAHH147 pKa = 6.92RR148 pKa = 11.84PWQMKK153 pKa = 9.77DD154 pKa = 2.99LQAIKK159 pKa = 10.78QEE161 pKa = 4.53VNTSAPGSPQFMQTVRR177 pKa = 11.84LAIQQFDD184 pKa = 3.85PTAKK188 pKa = 10.47DD189 pKa = 3.57LQDD192 pKa = 3.86LLQYY196 pKa = 10.43LCSSLVVSLHH206 pKa = 5.5HH207 pKa = 5.89QQLHH211 pKa = 4.61TLITEE216 pKa = 4.28AEE218 pKa = 4.24TRR220 pKa = 11.84GMTGYY225 pKa = 11.08NPMAGPLRR233 pKa = 11.84MQANNPAQEE242 pKa = 4.26GLRR245 pKa = 11.84RR246 pKa = 11.84EE247 pKa = 4.29YY248 pKa = 11.06QNLWLAAFSALPGNTRR264 pKa = 11.84DD265 pKa = 4.02PSWAAILQGLEE276 pKa = 3.97EE277 pKa = 5.02PYY279 pKa = 10.54CALLEE284 pKa = 4.22RR285 pKa = 11.84LNVALDD291 pKa = 3.32NGLPEE296 pKa = 4.52GTPKK300 pKa = 10.76EE301 pKa = 4.23PILRR305 pKa = 11.84SLAYY309 pKa = 10.74SNANKK314 pKa = 10.2DD315 pKa = 3.9CQKK318 pKa = 10.69LAGSGPYY325 pKa = 9.91
Molecular weight: 35.85 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|O09243|O09243_9DELA Env polyprotein OS=Simian T-lymphotropic virus 2 OX=33748 GN=env PE=4 SV=1
QQ1 pKa = 7.44GLISPSCPRR10 pKa = 11.84PLPPSQKK17 pKa = 10.23VSDD20 pKa = 3.98TTVLGAGGQTSSQFKK35 pKa = 10.44LLRR38 pKa = 11.84SPLCVYY44 pKa = 10.73LPFRR48 pKa = 11.84KK49 pKa = 10.07APVTLPSCLLDD60 pKa = 3.97TDD62 pKa = 4.34NKK64 pKa = 8.68WAIIGRR70 pKa = 11.84DD71 pKa = 3.26ILQQCQSVLYY81 pKa = 10.39LPEE84 pKa = 6.13DD85 pKa = 3.8NLCEE89 pKa = 4.16GTPRR93 pKa = 11.84PSGRR97 pKa = 11.84MNSPRR102 pKa = 11.84LLPVATPSVIGLEE115 pKa = 4.32HH116 pKa = 6.97FPPPPQIDD124 pKa = 3.41QFPFKK129 pKa = 10.45PEE131 pKa = 3.61RR132 pKa = 11.84LQALTDD138 pKa = 3.97LVSKK142 pKa = 10.65ALEE145 pKa = 3.79ASYY148 pKa = 10.52IEE150 pKa = 5.24PYY152 pKa = 10.2SGPGNNPVFPVKK164 pKa = 10.43KK165 pKa = 10.1PNGKK169 pKa = 8.28WRR171 pKa = 11.84FIHH174 pKa = 7.35DD175 pKa = 4.17LRR177 pKa = 11.84ATNAITTTLASPSPGPPDD195 pKa = 3.56LTSLSTALPYY205 pKa = 10.7VQTIDD210 pKa = 3.54LADD213 pKa = 3.89AFFQIPLPKK222 pKa = 10.09QFQPYY227 pKa = 9.21FAFTIPQPCNYY238 pKa = 10.01GPGARR243 pKa = 11.84YY244 pKa = 9.04AWTVLPQGFKK254 pKa = 10.78NSPTLFEE261 pKa = 4.06QQLAAILSPIRR272 pKa = 11.84KK273 pKa = 8.93AFPTSTIIQYY283 pKa = 9.66MDD285 pKa = 5.98DD286 pKa = 3.96ILLASPAQGEE296 pKa = 4.21LQQLSKK302 pKa = 8.45MTLQALVTHH311 pKa = 6.77GLPVSQAPRR320 pKa = 11.84EE321 pKa = 4.08QTPGQIRR328 pKa = 11.84FLGQVISPDD337 pKa = 3.45HH338 pKa = 5.92ITYY341 pKa = 7.62EE342 pKa = 4.26TTPTIPMKK350 pKa = 10.59SQWTLAEE357 pKa = 4.11LQTVLGEE364 pKa = 4.19IQWVSKK370 pKa = 8.15GTPILRR376 pKa = 11.84KK377 pKa = 9.41HH378 pKa = 5.63LQCLYY383 pKa = 10.28SALRR387 pKa = 11.84GYY389 pKa = 10.48QDD391 pKa = 3.32PRR393 pKa = 11.84AHH395 pKa = 7.42LLLQKK400 pKa = 10.18QQLHH404 pKa = 6.52ALHH407 pKa = 7.52AIQPLQHH414 pKa = 6.2NCRR417 pKa = 11.84SRR419 pKa = 11.84LNPALPILGLISLSSSGTTSVLFQARR445 pKa = 11.84QRR447 pKa = 11.84WPLVWLHH454 pKa = 6.04TPHH457 pKa = 7.45PPTSLGPWGHH467 pKa = 7.24LLACTILTLDD477 pKa = 4.04KK478 pKa = 11.3YY479 pKa = 11.05SLQHH483 pKa = 6.15YY484 pKa = 9.66GRR486 pKa = 11.84LCQSLEE492 pKa = 4.17DD493 pKa = 3.93NMSNTALHH501 pKa = 7.02DD502 pKa = 3.91FVKK505 pKa = 10.62NSPHH509 pKa = 6.08PRR511 pKa = 11.84VGILIHH517 pKa = 6.41HH518 pKa = 7.02MSRR521 pKa = 11.84FHH523 pKa = 6.88NLGSQPSGPWKK534 pKa = 10.69ALLHH538 pKa = 6.35LPALLQAARR547 pKa = 11.84LLRR550 pKa = 11.84PLFTLSPVVLTTHH563 pKa = 7.05PVSSPTGLPKK573 pKa = 10.21KK574 pKa = 10.01AAYY577 pKa = 9.5VLWDD581 pKa = 3.46QTILRR586 pKa = 11.84HH587 pKa = 6.97DD588 pKa = 5.07SITLPPHH595 pKa = 6.67GSNSAQRR602 pKa = 11.84GEE604 pKa = 4.18LLALLSGLRR613 pKa = 11.84AAKK616 pKa = 9.7SWPSLNIFLDD626 pKa = 3.54SKK628 pKa = 11.12YY629 pKa = 10.38LIKK632 pKa = 10.63YY633 pKa = 7.54LHH635 pKa = 6.54SLATGAFLGTSTHH648 pKa = 5.47QSLYY652 pKa = 10.36AHH654 pKa = 7.08LPALLHH660 pKa = 5.97NKK662 pKa = 9.34VIYY665 pKa = 9.47LHH667 pKa = 7.05HH668 pKa = 6.67IRR670 pKa = 11.84SHH672 pKa = 6.09TNLPDD677 pKa = 5.11PISTLNEE684 pKa = 4.05YY685 pKa = 9.62TDD687 pKa = 3.69SLIIIAPLIPLTPQDD702 pKa = 3.78LHH704 pKa = 7.61RR705 pKa = 11.84LTHH708 pKa = 6.11CNSRR712 pKa = 11.84GPCFFRR718 pKa = 11.84ATPQQSQVALEE729 pKa = 4.23SRR731 pKa = 11.84VTPATSLNSQHH742 pKa = 6.59HH743 pKa = 6.0MPQGHH748 pKa = 6.55IRR750 pKa = 11.84RR751 pKa = 11.84GLLPNHH757 pKa = 6.73IWQGDD762 pKa = 3.75VTHH765 pKa = 6.32YY766 pKa = 10.04KK767 pKa = 9.96YY768 pKa = 10.41KK769 pKa = 9.06RR770 pKa = 11.84HH771 pKa = 6.41RR772 pKa = 11.84YY773 pKa = 8.24CLHH776 pKa = 5.38VWVDD780 pKa = 3.51TFSNAVSITCKK791 pKa = 9.99TKK793 pKa = 8.25EE794 pKa = 4.21TSSEE798 pKa = 4.24TVSALLHH805 pKa = 6.81AITILGKK812 pKa = 9.44PLSINTDD819 pKa = 2.97NGSAFLSQEE828 pKa = 3.67FQAFCTSWHH837 pKa = 6.34IRR839 pKa = 11.84HH840 pKa = 5.76STHH843 pKa = 5.85VPYY846 pKa = 10.98NPTSSGLVEE855 pKa = 4.04RR856 pKa = 11.84TNGIVKK862 pKa = 10.48ALLNKK867 pKa = 10.2YY868 pKa = 10.52LLDD871 pKa = 4.37SPNLPLDD878 pKa = 3.59NAISKK883 pKa = 9.28SLWTLNQLNVMSPSGKK899 pKa = 8.01TRR901 pKa = 11.84WQLHH905 pKa = 5.66HH906 pKa = 7.01GPRR909 pKa = 11.84LPPSLQIPQPSKK921 pKa = 11.32APANWYY927 pKa = 9.69YY928 pKa = 11.52VLTPGLTNQRR938 pKa = 11.84WKK940 pKa = 11.29GPLHH944 pKa = 7.12LSRR947 pKa = 11.84KK948 pKa = 7.84LQEE951 pKa = 4.2RR952 pKa = 11.84LLSIDD957 pKa = 5.54GSPQWIPWRR966 pKa = 11.84LLKK969 pKa = 9.93KK970 pKa = 7.73TVCPRR975 pKa = 11.84PDD977 pKa = 3.48GSEE980 pKa = 4.06LAAHH984 pKa = 7.48AATDD988 pKa = 4.02HH989 pKa = 5.13QHH991 pKa = 6.46HH992 pKa = 6.57GG993 pKa = 3.58
QQ1 pKa = 7.44GLISPSCPRR10 pKa = 11.84PLPPSQKK17 pKa = 10.23VSDD20 pKa = 3.98TTVLGAGGQTSSQFKK35 pKa = 10.44LLRR38 pKa = 11.84SPLCVYY44 pKa = 10.73LPFRR48 pKa = 11.84KK49 pKa = 10.07APVTLPSCLLDD60 pKa = 3.97TDD62 pKa = 4.34NKK64 pKa = 8.68WAIIGRR70 pKa = 11.84DD71 pKa = 3.26ILQQCQSVLYY81 pKa = 10.39LPEE84 pKa = 6.13DD85 pKa = 3.8NLCEE89 pKa = 4.16GTPRR93 pKa = 11.84PSGRR97 pKa = 11.84MNSPRR102 pKa = 11.84LLPVATPSVIGLEE115 pKa = 4.32HH116 pKa = 6.97FPPPPQIDD124 pKa = 3.41QFPFKK129 pKa = 10.45PEE131 pKa = 3.61RR132 pKa = 11.84LQALTDD138 pKa = 3.97LVSKK142 pKa = 10.65ALEE145 pKa = 3.79ASYY148 pKa = 10.52IEE150 pKa = 5.24PYY152 pKa = 10.2SGPGNNPVFPVKK164 pKa = 10.43KK165 pKa = 10.1PNGKK169 pKa = 8.28WRR171 pKa = 11.84FIHH174 pKa = 7.35DD175 pKa = 4.17LRR177 pKa = 11.84ATNAITTTLASPSPGPPDD195 pKa = 3.56LTSLSTALPYY205 pKa = 10.7VQTIDD210 pKa = 3.54LADD213 pKa = 3.89AFFQIPLPKK222 pKa = 10.09QFQPYY227 pKa = 9.21FAFTIPQPCNYY238 pKa = 10.01GPGARR243 pKa = 11.84YY244 pKa = 9.04AWTVLPQGFKK254 pKa = 10.78NSPTLFEE261 pKa = 4.06QQLAAILSPIRR272 pKa = 11.84KK273 pKa = 8.93AFPTSTIIQYY283 pKa = 9.66MDD285 pKa = 5.98DD286 pKa = 3.96ILLASPAQGEE296 pKa = 4.21LQQLSKK302 pKa = 8.45MTLQALVTHH311 pKa = 6.77GLPVSQAPRR320 pKa = 11.84EE321 pKa = 4.08QTPGQIRR328 pKa = 11.84FLGQVISPDD337 pKa = 3.45HH338 pKa = 5.92ITYY341 pKa = 7.62EE342 pKa = 4.26TTPTIPMKK350 pKa = 10.59SQWTLAEE357 pKa = 4.11LQTVLGEE364 pKa = 4.19IQWVSKK370 pKa = 8.15GTPILRR376 pKa = 11.84KK377 pKa = 9.41HH378 pKa = 5.63LQCLYY383 pKa = 10.28SALRR387 pKa = 11.84GYY389 pKa = 10.48QDD391 pKa = 3.32PRR393 pKa = 11.84AHH395 pKa = 7.42LLLQKK400 pKa = 10.18QQLHH404 pKa = 6.52ALHH407 pKa = 7.52AIQPLQHH414 pKa = 6.2NCRR417 pKa = 11.84SRR419 pKa = 11.84LNPALPILGLISLSSSGTTSVLFQARR445 pKa = 11.84QRR447 pKa = 11.84WPLVWLHH454 pKa = 6.04TPHH457 pKa = 7.45PPTSLGPWGHH467 pKa = 7.24LLACTILTLDD477 pKa = 4.04KK478 pKa = 11.3YY479 pKa = 11.05SLQHH483 pKa = 6.15YY484 pKa = 9.66GRR486 pKa = 11.84LCQSLEE492 pKa = 4.17DD493 pKa = 3.93NMSNTALHH501 pKa = 7.02DD502 pKa = 3.91FVKK505 pKa = 10.62NSPHH509 pKa = 6.08PRR511 pKa = 11.84VGILIHH517 pKa = 6.41HH518 pKa = 7.02MSRR521 pKa = 11.84FHH523 pKa = 6.88NLGSQPSGPWKK534 pKa = 10.69ALLHH538 pKa = 6.35LPALLQAARR547 pKa = 11.84LLRR550 pKa = 11.84PLFTLSPVVLTTHH563 pKa = 7.05PVSSPTGLPKK573 pKa = 10.21KK574 pKa = 10.01AAYY577 pKa = 9.5VLWDD581 pKa = 3.46QTILRR586 pKa = 11.84HH587 pKa = 6.97DD588 pKa = 5.07SITLPPHH595 pKa = 6.67GSNSAQRR602 pKa = 11.84GEE604 pKa = 4.18LLALLSGLRR613 pKa = 11.84AAKK616 pKa = 9.7SWPSLNIFLDD626 pKa = 3.54SKK628 pKa = 11.12YY629 pKa = 10.38LIKK632 pKa = 10.63YY633 pKa = 7.54LHH635 pKa = 6.54SLATGAFLGTSTHH648 pKa = 5.47QSLYY652 pKa = 10.36AHH654 pKa = 7.08LPALLHH660 pKa = 5.97NKK662 pKa = 9.34VIYY665 pKa = 9.47LHH667 pKa = 7.05HH668 pKa = 6.67IRR670 pKa = 11.84SHH672 pKa = 6.09TNLPDD677 pKa = 5.11PISTLNEE684 pKa = 4.05YY685 pKa = 9.62TDD687 pKa = 3.69SLIIIAPLIPLTPQDD702 pKa = 3.78LHH704 pKa = 7.61RR705 pKa = 11.84LTHH708 pKa = 6.11CNSRR712 pKa = 11.84GPCFFRR718 pKa = 11.84ATPQQSQVALEE729 pKa = 4.23SRR731 pKa = 11.84VTPATSLNSQHH742 pKa = 6.59HH743 pKa = 6.0MPQGHH748 pKa = 6.55IRR750 pKa = 11.84RR751 pKa = 11.84GLLPNHH757 pKa = 6.73IWQGDD762 pKa = 3.75VTHH765 pKa = 6.32YY766 pKa = 10.04KK767 pKa = 9.96YY768 pKa = 10.41KK769 pKa = 9.06RR770 pKa = 11.84HH771 pKa = 6.41RR772 pKa = 11.84YY773 pKa = 8.24CLHH776 pKa = 5.38VWVDD780 pKa = 3.51TFSNAVSITCKK791 pKa = 9.99TKK793 pKa = 8.25EE794 pKa = 4.21TSSEE798 pKa = 4.24TVSALLHH805 pKa = 6.81AITILGKK812 pKa = 9.44PLSINTDD819 pKa = 2.97NGSAFLSQEE828 pKa = 3.67FQAFCTSWHH837 pKa = 6.34IRR839 pKa = 11.84HH840 pKa = 5.76STHH843 pKa = 5.85VPYY846 pKa = 10.98NPTSSGLVEE855 pKa = 4.04RR856 pKa = 11.84TNGIVKK862 pKa = 10.48ALLNKK867 pKa = 10.2YY868 pKa = 10.52LLDD871 pKa = 4.37SPNLPLDD878 pKa = 3.59NAISKK883 pKa = 9.28SLWTLNQLNVMSPSGKK899 pKa = 8.01TRR901 pKa = 11.84WQLHH905 pKa = 5.66HH906 pKa = 7.01GPRR909 pKa = 11.84LPPSLQIPQPSKK921 pKa = 11.32APANWYY927 pKa = 9.69YY928 pKa = 11.52VLTPGLTNQRR938 pKa = 11.84WKK940 pKa = 11.29GPLHH944 pKa = 7.12LSRR947 pKa = 11.84KK948 pKa = 7.84LQEE951 pKa = 4.2RR952 pKa = 11.84LLSIDD957 pKa = 5.54GSPQWIPWRR966 pKa = 11.84LLKK969 pKa = 9.93KK970 pKa = 7.73TVCPRR975 pKa = 11.84PDD977 pKa = 3.48GSEE980 pKa = 4.06LAAHH984 pKa = 7.48AATDD988 pKa = 4.02HH989 pKa = 5.13QHH991 pKa = 6.46HH992 pKa = 6.57GG993 pKa = 3.58
Molecular weight: 110.52 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1804 |
325 |
993 |
601.3 |
66.79 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.596 ± 0.601 | 2.051 ± 0.532 |
3.16 ± 0.045 | 2.55 ± 0.442 |
2.716 ± 0.141 | 5.155 ± 0.105 |
4.213 ± 0.517 | 5.266 ± 0.294 |
3.215 ± 0.659 | 14.024 ± 0.228 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
0.998 ± 0.216 | 3.88 ± 0.262 |
10.421 ± 0.975 | 6.375 ± 0.284 |
4.712 ± 0.024 | 9.091 ± 0.854 |
6.707 ± 0.485 | 3.769 ± 0.508 |
2.162 ± 0.268 | 2.938 ± 0.161 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
