
Lactobacillus bifermentans DSM 20003
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Lactobacillales; Lactobacillaceae; Loigolactobacillus; Loigolactobacillus bifermentans
Average proteome isoelectric point is 6.72
Get precalculated fractions of proteins
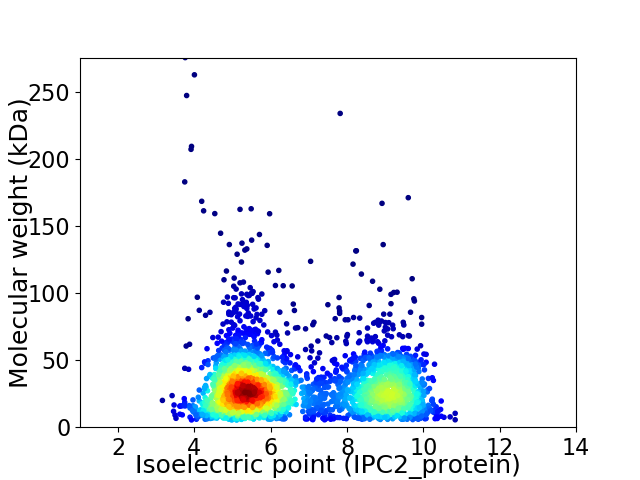
Virtual 2D-PAGE plot for 2964 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0R1H332|A0A0R1H332_9LACO Uncharacterized protein OS=Lactobacillus bifermentans DSM 20003 OX=1423726 GN=FC07_GL002564 PE=4 SV=1
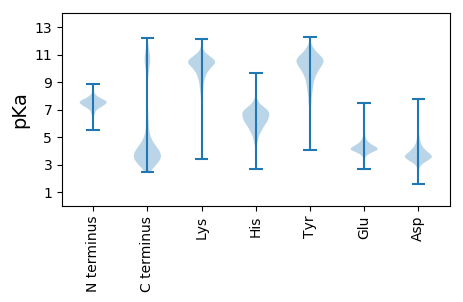
MM1 pKa = 7.74LDD3 pKa = 3.31EE4 pKa = 5.25NGFTRR9 pKa = 11.84PTYY12 pKa = 10.69DD13 pKa = 3.78EE14 pKa = 5.77LIQTLIQKK22 pKa = 7.83WQEE25 pKa = 3.88LFGEE29 pKa = 4.44NANVAPNSVGGIFIRR44 pKa = 11.84VLAFFLNQIYY54 pKa = 10.2QLAEE58 pKa = 3.77LVYY61 pKa = 10.19QSQFADD67 pKa = 3.44SATGTTLDD75 pKa = 3.74QLAANLGLVRR85 pKa = 11.84QAPQAAIGEE94 pKa = 4.27VQIYY98 pKa = 9.61GVAGYY103 pKa = 8.5VVPAGTLFQTDD114 pKa = 3.47DD115 pKa = 3.29GLIYY119 pKa = 9.82VTSEE123 pKa = 4.68DD124 pKa = 3.43ITLADD129 pKa = 3.53QGKK132 pKa = 7.59TSIDD136 pKa = 3.46TGDD139 pKa = 3.67LGQGVLQYY147 pKa = 10.93NDD149 pKa = 3.53KK150 pKa = 11.22NIGLGTSTVLYY161 pKa = 11.05ANGTGSNYY169 pKa = 10.29NKK171 pKa = 9.95PGVIGDD177 pKa = 4.37YY178 pKa = 10.25IAQQLMPVEE187 pKa = 4.23EE188 pKa = 4.23VLLVEE193 pKa = 4.34VGKK196 pKa = 8.85ITGGADD202 pKa = 4.36LEE204 pKa = 5.29DD205 pKa = 5.28DD206 pKa = 3.84DD207 pKa = 5.54ALRR210 pKa = 11.84DD211 pKa = 3.77RR212 pKa = 11.84LEE214 pKa = 4.09QASQEE219 pKa = 4.48APSSPYY225 pKa = 10.05NGVMSAVRR233 pKa = 11.84DD234 pKa = 3.88VVGVSSAKK242 pKa = 10.18IVVNDD247 pKa = 4.26TMDD250 pKa = 3.41TDD252 pKa = 4.46ASGNPAKK259 pKa = 9.7TLHH262 pKa = 6.57IYY264 pKa = 10.6VDD266 pKa = 4.27GGNQDD271 pKa = 4.85DD272 pKa = 3.72IGAAIFDD279 pKa = 4.66SIAAGIQTYY288 pKa = 10.64GSIEE292 pKa = 4.4VYY294 pKa = 10.73VKK296 pKa = 10.63DD297 pKa = 3.76IGGSTHH303 pKa = 6.06SVYY306 pKa = 10.8YY307 pKa = 9.74DD308 pKa = 3.28QPTAIQIFASVAATTNEE325 pKa = 3.58AFPLDD330 pKa = 4.04GNAQIQQAVVDD341 pKa = 4.26YY342 pKa = 10.16VRR344 pKa = 11.84SVGMGGTIHH353 pKa = 6.74YY354 pKa = 10.05SYY356 pKa = 10.64LYY358 pKa = 10.29KK359 pKa = 10.66YY360 pKa = 10.48LYY362 pKa = 10.97DD363 pKa = 4.84NITGLDD369 pKa = 3.54VADD372 pKa = 4.0VKK374 pKa = 11.0IGTDD378 pKa = 3.43KK379 pKa = 11.68DD380 pKa = 3.5NLTAADD386 pKa = 4.38IPLTDD391 pKa = 3.64IQRR394 pKa = 11.84ATITTDD400 pKa = 3.26TVVVAA405 pKa = 5.14
MM1 pKa = 7.74LDD3 pKa = 3.31EE4 pKa = 5.25NGFTRR9 pKa = 11.84PTYY12 pKa = 10.69DD13 pKa = 3.78EE14 pKa = 5.77LIQTLIQKK22 pKa = 7.83WQEE25 pKa = 3.88LFGEE29 pKa = 4.44NANVAPNSVGGIFIRR44 pKa = 11.84VLAFFLNQIYY54 pKa = 10.2QLAEE58 pKa = 3.77LVYY61 pKa = 10.19QSQFADD67 pKa = 3.44SATGTTLDD75 pKa = 3.74QLAANLGLVRR85 pKa = 11.84QAPQAAIGEE94 pKa = 4.27VQIYY98 pKa = 9.61GVAGYY103 pKa = 8.5VVPAGTLFQTDD114 pKa = 3.47DD115 pKa = 3.29GLIYY119 pKa = 9.82VTSEE123 pKa = 4.68DD124 pKa = 3.43ITLADD129 pKa = 3.53QGKK132 pKa = 7.59TSIDD136 pKa = 3.46TGDD139 pKa = 3.67LGQGVLQYY147 pKa = 10.93NDD149 pKa = 3.53KK150 pKa = 11.22NIGLGTSTVLYY161 pKa = 11.05ANGTGSNYY169 pKa = 10.29NKK171 pKa = 9.95PGVIGDD177 pKa = 4.37YY178 pKa = 10.25IAQQLMPVEE187 pKa = 4.23EE188 pKa = 4.23VLLVEE193 pKa = 4.34VGKK196 pKa = 8.85ITGGADD202 pKa = 4.36LEE204 pKa = 5.29DD205 pKa = 5.28DD206 pKa = 3.84DD207 pKa = 5.54ALRR210 pKa = 11.84DD211 pKa = 3.77RR212 pKa = 11.84LEE214 pKa = 4.09QASQEE219 pKa = 4.48APSSPYY225 pKa = 10.05NGVMSAVRR233 pKa = 11.84DD234 pKa = 3.88VVGVSSAKK242 pKa = 10.18IVVNDD247 pKa = 4.26TMDD250 pKa = 3.41TDD252 pKa = 4.46ASGNPAKK259 pKa = 9.7TLHH262 pKa = 6.57IYY264 pKa = 10.6VDD266 pKa = 4.27GGNQDD271 pKa = 4.85DD272 pKa = 3.72IGAAIFDD279 pKa = 4.66SIAAGIQTYY288 pKa = 10.64GSIEE292 pKa = 4.4VYY294 pKa = 10.73VKK296 pKa = 10.63DD297 pKa = 3.76IGGSTHH303 pKa = 6.06SVYY306 pKa = 10.8YY307 pKa = 9.74DD308 pKa = 3.28QPTAIQIFASVAATTNEE325 pKa = 3.58AFPLDD330 pKa = 4.04GNAQIQQAVVDD341 pKa = 4.26YY342 pKa = 10.16VRR344 pKa = 11.84SVGMGGTIHH353 pKa = 6.74YY354 pKa = 10.05SYY356 pKa = 10.64LYY358 pKa = 10.29KK359 pKa = 10.66YY360 pKa = 10.48LYY362 pKa = 10.97DD363 pKa = 4.84NITGLDD369 pKa = 3.54VADD372 pKa = 4.0VKK374 pKa = 11.0IGTDD378 pKa = 3.43KK379 pKa = 11.68DD380 pKa = 3.5NLTAADD386 pKa = 4.38IPLTDD391 pKa = 3.64IQRR394 pKa = 11.84ATITTDD400 pKa = 3.26TVVVAA405 pKa = 5.14
Molecular weight: 43.17 kDa
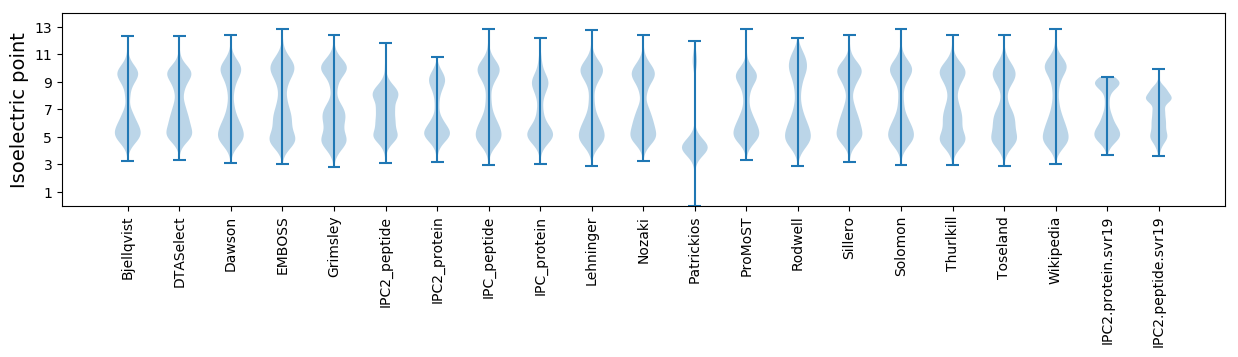
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0R1GQZ8|A0A0R1GQZ8_9LACO ABC transporter OS=Lactobacillus bifermentans DSM 20003 OX=1423726 GN=FC07_GL000823 PE=4 SV=1
MM1 pKa = 7.65HH2 pKa = 7.37YY3 pKa = 10.39IDD5 pKa = 3.64ITGQRR10 pKa = 11.84FGKK13 pKa = 10.34LVAQTCGSPDD23 pKa = 3.17RR24 pKa = 11.84QGNMLWHH31 pKa = 6.22CTCDD35 pKa = 2.94CGRR38 pKa = 11.84TLLVSGTNLRR48 pKa = 11.84QGRR51 pKa = 11.84QKK53 pKa = 11.08SCGQCGSLRR62 pKa = 11.84LIDD65 pKa = 3.6LTGQRR70 pKa = 11.84FGRR73 pKa = 11.84LVVMKK78 pKa = 10.47RR79 pKa = 11.84STQRR83 pKa = 11.84SANGNALWQCRR94 pKa = 11.84CDD96 pKa = 3.31CGKK99 pKa = 10.24QVVVDD104 pKa = 3.64SQRR107 pKa = 11.84LRR109 pKa = 11.84KK110 pKa = 9.81HH111 pKa = 4.66ITRR114 pKa = 11.84SCGCLRR120 pKa = 11.84NEE122 pKa = 4.09LAKK125 pKa = 10.54KK126 pKa = 10.42RR127 pKa = 11.84SYY129 pKa = 11.2HH130 pKa = 5.25NAAFRR135 pKa = 11.84KK136 pKa = 6.83TQGNISRR143 pKa = 11.84LKK145 pKa = 10.02DD146 pKa = 3.27ANGVFFCSTKK156 pKa = 8.27KK157 pKa = 8.96TKK159 pKa = 10.33RR160 pKa = 11.84NRR162 pKa = 11.84TGVIGVSFDD171 pKa = 3.42QHH173 pKa = 6.17SGRR176 pKa = 11.84YY177 pKa = 6.38VARR180 pKa = 11.84LRR182 pKa = 11.84YY183 pKa = 9.44RR184 pKa = 11.84GQYY187 pKa = 9.91VLNQTAATLQEE198 pKa = 4.39AATLRR203 pKa = 11.84HH204 pKa = 5.69RR205 pKa = 11.84AEE207 pKa = 3.97LKK209 pKa = 9.86YY210 pKa = 10.33FKK212 pKa = 10.48PSTTDD217 pKa = 2.65
MM1 pKa = 7.65HH2 pKa = 7.37YY3 pKa = 10.39IDD5 pKa = 3.64ITGQRR10 pKa = 11.84FGKK13 pKa = 10.34LVAQTCGSPDD23 pKa = 3.17RR24 pKa = 11.84QGNMLWHH31 pKa = 6.22CTCDD35 pKa = 2.94CGRR38 pKa = 11.84TLLVSGTNLRR48 pKa = 11.84QGRR51 pKa = 11.84QKK53 pKa = 11.08SCGQCGSLRR62 pKa = 11.84LIDD65 pKa = 3.6LTGQRR70 pKa = 11.84FGRR73 pKa = 11.84LVVMKK78 pKa = 10.47RR79 pKa = 11.84STQRR83 pKa = 11.84SANGNALWQCRR94 pKa = 11.84CDD96 pKa = 3.31CGKK99 pKa = 10.24QVVVDD104 pKa = 3.64SQRR107 pKa = 11.84LRR109 pKa = 11.84KK110 pKa = 9.81HH111 pKa = 4.66ITRR114 pKa = 11.84SCGCLRR120 pKa = 11.84NEE122 pKa = 4.09LAKK125 pKa = 10.54KK126 pKa = 10.42RR127 pKa = 11.84SYY129 pKa = 11.2HH130 pKa = 5.25NAAFRR135 pKa = 11.84KK136 pKa = 6.83TQGNISRR143 pKa = 11.84LKK145 pKa = 10.02DD146 pKa = 3.27ANGVFFCSTKK156 pKa = 8.27KK157 pKa = 8.96TKK159 pKa = 10.33RR160 pKa = 11.84NRR162 pKa = 11.84TGVIGVSFDD171 pKa = 3.42QHH173 pKa = 6.17SGRR176 pKa = 11.84YY177 pKa = 6.38VARR180 pKa = 11.84LRR182 pKa = 11.84YY183 pKa = 9.44RR184 pKa = 11.84GQYY187 pKa = 9.91VLNQTAATLQEE198 pKa = 4.39AATLRR203 pKa = 11.84HH204 pKa = 5.69RR205 pKa = 11.84AEE207 pKa = 3.97LKK209 pKa = 9.86YY210 pKa = 10.33FKK212 pKa = 10.48PSTTDD217 pKa = 2.65
Molecular weight: 24.54 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
885647 |
46 |
2691 |
298.8 |
33.06 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.813 ± 0.078 | 0.629 ± 0.015 |
5.313 ± 0.048 | 4.6 ± 0.046 |
4.081 ± 0.036 | 6.719 ± 0.044 |
2.291 ± 0.022 | 6.197 ± 0.047 |
5.283 ± 0.056 | 10.509 ± 0.072 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.471 ± 0.025 | 3.999 ± 0.032 |
3.988 ± 0.035 | 6.145 ± 0.073 |
4.125 ± 0.047 | 5.114 ± 0.05 |
7.112 ± 0.089 | 7.11 ± 0.043 |
1.098 ± 0.018 | 3.388 ± 0.027 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
