
Sewage-associated circular DNA virus-9
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cremevirales; Smacoviridae; unclassified Smacoviridae
Average proteome isoelectric point is 4.83
Get precalculated fractions of proteins
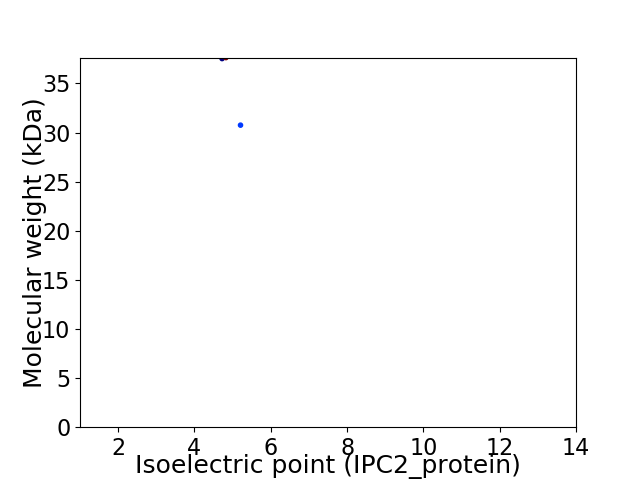
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A075IZU5|A0A075IZU5_9VIRU Replication-associated protein OS=Sewage-associated circular DNA virus-9 OX=1519398 PE=4 SV=1
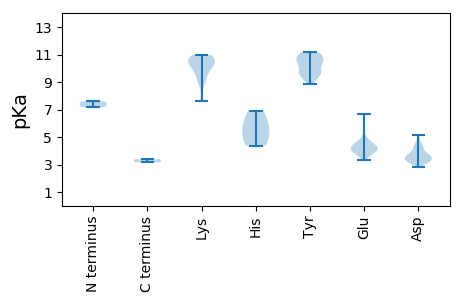
MM1 pKa = 7.17VFVKK5 pKa = 10.66VSEE8 pKa = 4.46TYY10 pKa = 10.74DD11 pKa = 3.11LSTNVGKK18 pKa = 9.83MGIVGIHH25 pKa = 5.33TPKK28 pKa = 10.77GDD30 pKa = 4.18LISRR34 pKa = 11.84LWGGLAMQHH43 pKa = 6.23KK44 pKa = 9.1YY45 pKa = 9.14VRR47 pKa = 11.84FDD49 pKa = 3.43SCDD52 pKa = 3.07VTMACASLLPADD64 pKa = 4.64PLQIGQEE71 pKa = 4.04AGSIAPQDD79 pKa = 3.62MFNPILYY86 pKa = 9.77KK87 pKa = 10.6AVSNDD92 pKa = 3.23SMSNLQAFLSATATRR107 pKa = 11.84DD108 pKa = 3.71SQIGTAKK115 pKa = 10.57GSVVDD120 pKa = 5.18LNDD123 pKa = 4.25PNFQFNDD130 pKa = 3.7SVPINQFDD138 pKa = 3.25IYY140 pKa = 11.11YY141 pKa = 10.13SLLGNPDD148 pKa = 3.42GFKK151 pKa = 10.78KK152 pKa = 10.64AMPQAGLQMRR162 pKa = 11.84GLYY165 pKa = 9.43PLVYY169 pKa = 9.87QVVSNYY175 pKa = 9.62GVNTAGMTDD184 pKa = 3.35SDD186 pKa = 5.03SIGNNSNPSNPIYY199 pKa = 10.96GPDD202 pKa = 3.57FQDD205 pKa = 3.35GVTPLDD211 pKa = 3.24ISNIQLRR218 pKa = 11.84GPAMRR223 pKa = 11.84MPRR226 pKa = 11.84VNTTYY231 pKa = 10.87LPYY234 pKa = 10.91NSIQNAPLSGKK245 pKa = 10.55ANLNFGDD252 pKa = 4.32TTRR255 pKa = 11.84GMPPCYY261 pKa = 9.6VAMIILPPATLNKK274 pKa = 10.11LYY276 pKa = 10.84YY277 pKa = 9.18RR278 pKa = 11.84LKK280 pKa = 8.98VTWTIEE286 pKa = 4.01FTEE289 pKa = 4.39LRR291 pKa = 11.84SLDD294 pKa = 4.5EE295 pKa = 4.02ISNWRR300 pKa = 11.84GMSEE304 pKa = 3.73IGVDD308 pKa = 4.48VYY310 pKa = 11.14ATDD313 pKa = 3.71YY314 pKa = 8.85ATQSAALMSDD324 pKa = 3.78MEE326 pKa = 4.83SMVDD330 pKa = 3.23VGNANIQKK338 pKa = 9.94IMEE341 pKa = 4.25GRR343 pKa = 3.39
MM1 pKa = 7.17VFVKK5 pKa = 10.66VSEE8 pKa = 4.46TYY10 pKa = 10.74DD11 pKa = 3.11LSTNVGKK18 pKa = 9.83MGIVGIHH25 pKa = 5.33TPKK28 pKa = 10.77GDD30 pKa = 4.18LISRR34 pKa = 11.84LWGGLAMQHH43 pKa = 6.23KK44 pKa = 9.1YY45 pKa = 9.14VRR47 pKa = 11.84FDD49 pKa = 3.43SCDD52 pKa = 3.07VTMACASLLPADD64 pKa = 4.64PLQIGQEE71 pKa = 4.04AGSIAPQDD79 pKa = 3.62MFNPILYY86 pKa = 9.77KK87 pKa = 10.6AVSNDD92 pKa = 3.23SMSNLQAFLSATATRR107 pKa = 11.84DD108 pKa = 3.71SQIGTAKK115 pKa = 10.57GSVVDD120 pKa = 5.18LNDD123 pKa = 4.25PNFQFNDD130 pKa = 3.7SVPINQFDD138 pKa = 3.25IYY140 pKa = 11.11YY141 pKa = 10.13SLLGNPDD148 pKa = 3.42GFKK151 pKa = 10.78KK152 pKa = 10.64AMPQAGLQMRR162 pKa = 11.84GLYY165 pKa = 9.43PLVYY169 pKa = 9.87QVVSNYY175 pKa = 9.62GVNTAGMTDD184 pKa = 3.35SDD186 pKa = 5.03SIGNNSNPSNPIYY199 pKa = 10.96GPDD202 pKa = 3.57FQDD205 pKa = 3.35GVTPLDD211 pKa = 3.24ISNIQLRR218 pKa = 11.84GPAMRR223 pKa = 11.84MPRR226 pKa = 11.84VNTTYY231 pKa = 10.87LPYY234 pKa = 10.91NSIQNAPLSGKK245 pKa = 10.55ANLNFGDD252 pKa = 4.32TTRR255 pKa = 11.84GMPPCYY261 pKa = 9.6VAMIILPPATLNKK274 pKa = 10.11LYY276 pKa = 10.84YY277 pKa = 9.18RR278 pKa = 11.84LKK280 pKa = 8.98VTWTIEE286 pKa = 4.01FTEE289 pKa = 4.39LRR291 pKa = 11.84SLDD294 pKa = 4.5EE295 pKa = 4.02ISNWRR300 pKa = 11.84GMSEE304 pKa = 3.73IGVDD308 pKa = 4.48VYY310 pKa = 11.14ATDD313 pKa = 3.71YY314 pKa = 8.85ATQSAALMSDD324 pKa = 3.78MEE326 pKa = 4.83SMVDD330 pKa = 3.23VGNANIQKK338 pKa = 9.94IMEE341 pKa = 4.25GRR343 pKa = 3.39
Molecular weight: 37.53 kDa
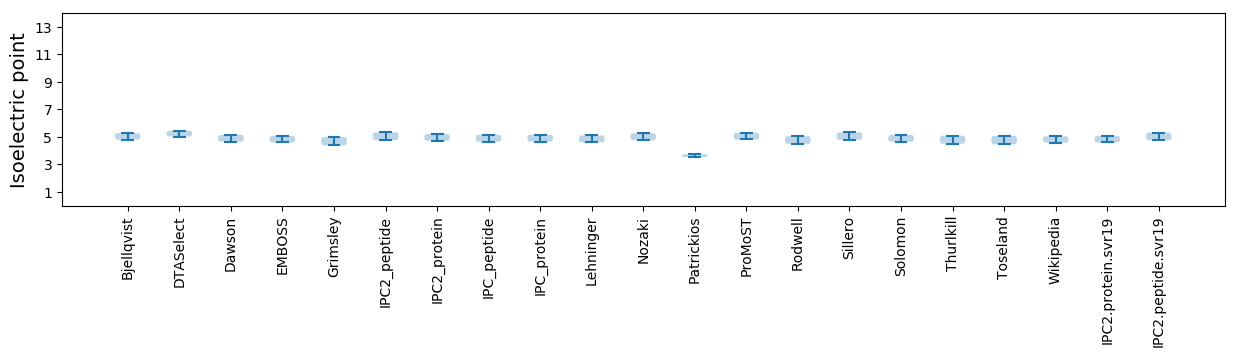
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A075IZU5|A0A075IZU5_9VIRU Replication-associated protein OS=Sewage-associated circular DNA virus-9 OX=1519398 PE=4 SV=1
MM1 pKa = 7.59ANVKK5 pKa = 9.67WIDD8 pKa = 3.29ATIWCEE14 pKa = 3.33GDD16 pKa = 3.2NAFEE20 pKa = 4.17GVKK23 pKa = 10.6DD24 pKa = 3.7EE25 pKa = 4.5SEE27 pKa = 3.79WRR29 pKa = 11.84SRR31 pKa = 11.84FAGLFEE37 pKa = 4.63RR38 pKa = 11.84YY39 pKa = 9.68AYY41 pKa = 10.25GRR43 pKa = 11.84EE44 pKa = 3.98TAPEE48 pKa = 3.63TGRR51 pKa = 11.84RR52 pKa = 11.84HH53 pKa = 4.37FQFRR57 pKa = 11.84GVLKK61 pKa = 9.28VACDD65 pKa = 4.15ASCLAYY71 pKa = 10.54LSSLGFRR78 pKa = 11.84NISPTHH84 pKa = 5.72VRR86 pKa = 11.84DD87 pKa = 3.2FDD89 pKa = 3.93YY90 pKa = 11.16VYY92 pKa = 11.01KK93 pKa = 10.98DD94 pKa = 3.39RR95 pKa = 11.84DD96 pKa = 4.16FFCSWDD102 pKa = 3.66VYY104 pKa = 10.45RR105 pKa = 11.84PEE107 pKa = 4.17YY108 pKa = 10.9DD109 pKa = 3.23LVRR112 pKa = 11.84NSPHH116 pKa = 4.7VWQVQLEE123 pKa = 4.14DD124 pKa = 3.68MEE126 pKa = 6.7RR127 pKa = 11.84DD128 pKa = 3.32DD129 pKa = 4.0RR130 pKa = 11.84TIEE133 pKa = 4.08IVWDD137 pKa = 3.43EE138 pKa = 4.43RR139 pKa = 11.84GNSGKK144 pKa = 7.63TAWAMYY150 pKa = 10.23QDD152 pKa = 4.4YY153 pKa = 10.37LHH155 pKa = 6.9KK156 pKa = 10.36AVYY159 pKa = 9.78IPPPQRR165 pKa = 11.84GLDD168 pKa = 3.56LVACVLGKK176 pKa = 10.23RR177 pKa = 11.84EE178 pKa = 4.14SDD180 pKa = 3.16WYY182 pKa = 10.45IIDD185 pKa = 3.59TPRR188 pKa = 11.84AFEE191 pKa = 4.3FTDD194 pKa = 3.67DD195 pKa = 3.64WACSIEE201 pKa = 3.98QLKK204 pKa = 10.52NGYY207 pKa = 9.48VFDD210 pKa = 3.62TRR212 pKa = 11.84YY213 pKa = 10.29SFRR216 pKa = 11.84DD217 pKa = 3.26RR218 pKa = 11.84YY219 pKa = 10.56LSVRR223 pKa = 11.84PRR225 pKa = 11.84VTVLCNTLPEE235 pKa = 4.06YY236 pKa = 10.58EE237 pKa = 4.87RR238 pKa = 11.84YY239 pKa = 9.74FSADD243 pKa = 2.79RR244 pKa = 11.84VLPFRR249 pKa = 11.84ITPEE253 pKa = 4.44GYY255 pKa = 9.44LWSVV259 pKa = 3.16
MM1 pKa = 7.59ANVKK5 pKa = 9.67WIDD8 pKa = 3.29ATIWCEE14 pKa = 3.33GDD16 pKa = 3.2NAFEE20 pKa = 4.17GVKK23 pKa = 10.6DD24 pKa = 3.7EE25 pKa = 4.5SEE27 pKa = 3.79WRR29 pKa = 11.84SRR31 pKa = 11.84FAGLFEE37 pKa = 4.63RR38 pKa = 11.84YY39 pKa = 9.68AYY41 pKa = 10.25GRR43 pKa = 11.84EE44 pKa = 3.98TAPEE48 pKa = 3.63TGRR51 pKa = 11.84RR52 pKa = 11.84HH53 pKa = 4.37FQFRR57 pKa = 11.84GVLKK61 pKa = 9.28VACDD65 pKa = 4.15ASCLAYY71 pKa = 10.54LSSLGFRR78 pKa = 11.84NISPTHH84 pKa = 5.72VRR86 pKa = 11.84DD87 pKa = 3.2FDD89 pKa = 3.93YY90 pKa = 11.16VYY92 pKa = 11.01KK93 pKa = 10.98DD94 pKa = 3.39RR95 pKa = 11.84DD96 pKa = 4.16FFCSWDD102 pKa = 3.66VYY104 pKa = 10.45RR105 pKa = 11.84PEE107 pKa = 4.17YY108 pKa = 10.9DD109 pKa = 3.23LVRR112 pKa = 11.84NSPHH116 pKa = 4.7VWQVQLEE123 pKa = 4.14DD124 pKa = 3.68MEE126 pKa = 6.7RR127 pKa = 11.84DD128 pKa = 3.32DD129 pKa = 4.0RR130 pKa = 11.84TIEE133 pKa = 4.08IVWDD137 pKa = 3.43EE138 pKa = 4.43RR139 pKa = 11.84GNSGKK144 pKa = 7.63TAWAMYY150 pKa = 10.23QDD152 pKa = 4.4YY153 pKa = 10.37LHH155 pKa = 6.9KK156 pKa = 10.36AVYY159 pKa = 9.78IPPPQRR165 pKa = 11.84GLDD168 pKa = 3.56LVACVLGKK176 pKa = 10.23RR177 pKa = 11.84EE178 pKa = 4.14SDD180 pKa = 3.16WYY182 pKa = 10.45IIDD185 pKa = 3.59TPRR188 pKa = 11.84AFEE191 pKa = 4.3FTDD194 pKa = 3.67DD195 pKa = 3.64WACSIEE201 pKa = 3.98QLKK204 pKa = 10.52NGYY207 pKa = 9.48VFDD210 pKa = 3.62TRR212 pKa = 11.84YY213 pKa = 10.29SFRR216 pKa = 11.84DD217 pKa = 3.26RR218 pKa = 11.84YY219 pKa = 10.56LSVRR223 pKa = 11.84PRR225 pKa = 11.84VTVLCNTLPEE235 pKa = 4.06YY236 pKa = 10.58EE237 pKa = 4.87RR238 pKa = 11.84YY239 pKa = 9.74FSADD243 pKa = 2.79RR244 pKa = 11.84VLPFRR249 pKa = 11.84ITPEE253 pKa = 4.44GYY255 pKa = 9.44LWSVV259 pKa = 3.16
Molecular weight: 30.75 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
602 |
259 |
343 |
301.0 |
34.14 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.645 ± 0.281 | 1.661 ± 0.627 |
7.807 ± 0.645 | 4.319 ± 1.583 |
4.319 ± 0.886 | 6.645 ± 0.978 |
0.997 ± 0.329 | 5.15 ± 0.775 |
3.322 ± 0.14 | 7.475 ± 0.548 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.488 ± 1.402 | 5.316 ± 1.572 |
5.648 ± 0.61 | 3.654 ± 0.805 |
6.312 ± 2.242 | 7.143 ± 0.813 |
5.316 ± 0.411 | 7.143 ± 0.348 |
2.159 ± 1.024 | 5.482 ± 0.651 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
