
Streptococcus phage Javan577
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Siphoviridae; unclassified Siphoviridae
Average proteome isoelectric point is 6.46
Get precalculated fractions of proteins
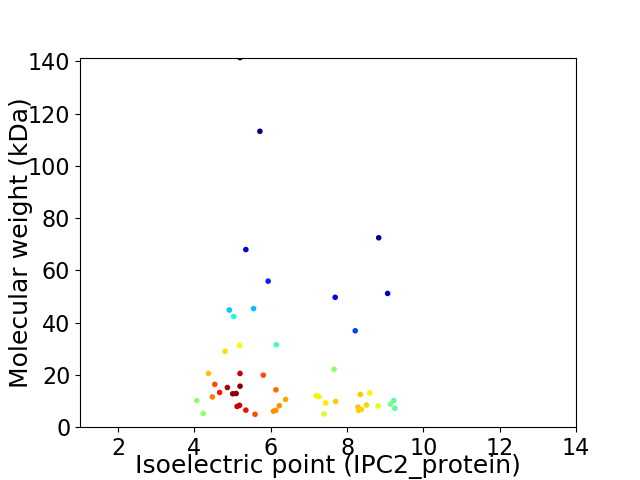
Virtual 2D-PAGE plot for 51 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
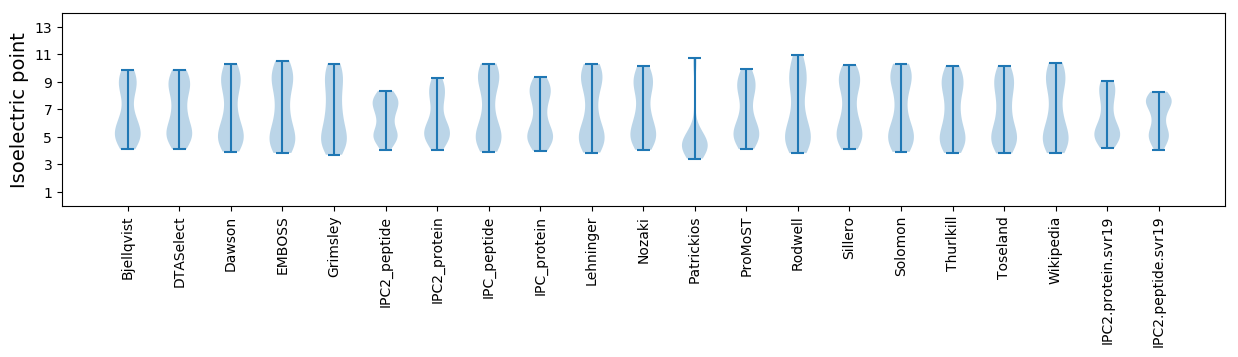
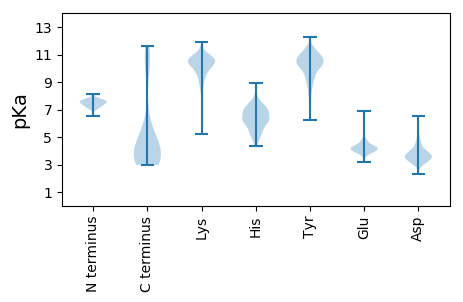
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4D6AKQ9|A0A4D6AKQ9_9CAUD Terminase large subunit OS=Streptococcus phage Javan577 OX=2548269 GN=Javan577_0029 PE=4 SV=1
MM1 pKa = 7.33QSTGLFSTFSEE12 pKa = 4.69DD13 pKa = 4.38EE14 pKa = 4.15LFEE17 pKa = 6.72DD18 pKa = 6.13DD19 pKa = 4.96VIFWTYY25 pKa = 10.62FDD27 pKa = 4.26EE28 pKa = 6.49FEE30 pKa = 4.45DD31 pKa = 3.54TGKK34 pKa = 10.74ARR36 pKa = 11.84IVHH39 pKa = 7.19RR40 pKa = 11.84DD41 pKa = 3.64GCWKK45 pKa = 10.53LLDD48 pKa = 3.43IKK50 pKa = 10.73AGKK53 pKa = 8.71EE54 pKa = 3.89VWDD57 pKa = 3.96SLFDD61 pKa = 3.83CLEE64 pKa = 4.04NCTVFLSGNIYY75 pKa = 10.69EE76 pKa = 4.55NLEE79 pKa = 3.98LMEE82 pKa = 4.58GLNEE86 pKa = 4.14KK87 pKa = 10.34
MM1 pKa = 7.33QSTGLFSTFSEE12 pKa = 4.69DD13 pKa = 4.38EE14 pKa = 4.15LFEE17 pKa = 6.72DD18 pKa = 6.13DD19 pKa = 4.96VIFWTYY25 pKa = 10.62FDD27 pKa = 4.26EE28 pKa = 6.49FEE30 pKa = 4.45DD31 pKa = 3.54TGKK34 pKa = 10.74ARR36 pKa = 11.84IVHH39 pKa = 7.19RR40 pKa = 11.84DD41 pKa = 3.64GCWKK45 pKa = 10.53LLDD48 pKa = 3.43IKK50 pKa = 10.73AGKK53 pKa = 8.71EE54 pKa = 3.89VWDD57 pKa = 3.96SLFDD61 pKa = 3.83CLEE64 pKa = 4.04NCTVFLSGNIYY75 pKa = 10.69EE76 pKa = 4.55NLEE79 pKa = 3.98LMEE82 pKa = 4.58GLNEE86 pKa = 4.14KK87 pKa = 10.34
Molecular weight: 10.19 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4D6ALG6|A0A4D6ALG6_9CAUD Uncharacterized protein OS=Streptococcus phage Javan577 OX=2548269 GN=Javan577_0011 PE=4 SV=1
MM1 pKa = 7.62KK2 pKa = 9.87EE3 pKa = 3.74HH4 pKa = 6.99SIKK7 pKa = 10.6KK8 pKa = 9.92LLTGTITLLSVVTLVACSQSNKK30 pKa = 10.41GIIDD34 pKa = 3.68CRR36 pKa = 11.84IKK38 pKa = 10.68KK39 pKa = 10.47SPTLNFGRR47 pKa = 11.84SEE49 pKa = 3.91RR50 pKa = 11.84KK51 pKa = 9.59ANHH54 pKa = 5.48VWSEE58 pKa = 4.02TCFAVGLFTIPIYY71 pKa = 9.47QKK73 pKa = 10.83RR74 pKa = 11.84KK75 pKa = 7.48GKK77 pKa = 10.54SMAYY81 pKa = 8.96FRR83 pKa = 11.84KK84 pKa = 9.89RR85 pKa = 11.84EE86 pKa = 4.06NGWEE90 pKa = 3.66YY91 pKa = 10.51RR92 pKa = 11.84ISYY95 pKa = 8.24KK96 pKa = 10.93APDD99 pKa = 3.57GSYY102 pKa = 9.91KK103 pKa = 10.36QKK105 pKa = 10.58SKK107 pKa = 11.15SGFRR111 pKa = 11.84TKK113 pKa = 10.73SEE115 pKa = 3.93AVQAASQAEE124 pKa = 4.22IEE126 pKa = 4.31LSSGIVEE133 pKa = 4.78DD134 pKa = 4.43KK135 pKa = 10.91NISLAEE141 pKa = 3.97YY142 pKa = 9.35FEE144 pKa = 4.32KK145 pKa = 10.39WMEE148 pKa = 3.84IHH150 pKa = 6.86KK151 pKa = 10.21KK152 pKa = 9.28PHH154 pKa = 6.29VDD156 pKa = 2.98PEE158 pKa = 4.45TYY160 pKa = 9.81RR161 pKa = 11.84KK162 pKa = 10.35YY163 pKa = 10.95EE164 pKa = 4.03FNLKK168 pKa = 9.81IIKK171 pKa = 9.76EE172 pKa = 4.23YY173 pKa = 9.84YY174 pKa = 10.29QNTKK178 pKa = 10.02LSKK181 pKa = 9.44ITTTMHH187 pKa = 4.98QQVLNKK193 pKa = 10.34LADD196 pKa = 3.98RR197 pKa = 11.84YY198 pKa = 10.75VKK200 pKa = 10.0YY201 pKa = 9.04TVKK204 pKa = 10.61LFNSHH209 pKa = 4.62IRR211 pKa = 11.84AAIKK215 pKa = 10.31VALHH219 pKa = 5.79QGTLKK224 pKa = 10.66KK225 pKa = 10.75DD226 pKa = 3.45FTQLAKK232 pKa = 10.7VFSNVEE238 pKa = 4.17SKK240 pKa = 11.16DD241 pKa = 3.48EE242 pKa = 4.13SEE244 pKa = 6.13KK245 pKa = 10.63YY246 pKa = 10.66LEE248 pKa = 4.3LNGYY252 pKa = 9.96RR253 pKa = 11.84SLLLDD258 pKa = 3.17VRR260 pKa = 11.84QSINHH265 pKa = 5.28QSHH268 pKa = 6.24FFIYY272 pKa = 9.97TIGKK276 pKa = 6.89TGLRR280 pKa = 11.84FSEE283 pKa = 4.07ATGLSKK289 pKa = 10.0FTVDD293 pKa = 3.28RR294 pKa = 11.84QNMCLHH300 pKa = 5.75IRR302 pKa = 11.84KK303 pKa = 7.75TYY305 pKa = 10.1KK306 pKa = 10.6VYY308 pKa = 10.59GKK310 pKa = 10.35KK311 pKa = 10.25KK312 pKa = 9.78GWGPTKK318 pKa = 10.55NPQSEE323 pKa = 3.99RR324 pKa = 11.84DD325 pKa = 3.66VPIDD329 pKa = 3.87RR330 pKa = 11.84DD331 pKa = 3.21WLEE334 pKa = 4.8AYY336 pKa = 10.11DD337 pKa = 4.26EE338 pKa = 4.21YY339 pKa = 11.35MEE341 pKa = 5.0VGYY344 pKa = 10.28IDD346 pKa = 4.5NPEE349 pKa = 3.55KK350 pKa = 10.8RR351 pKa = 11.84LFFKK355 pKa = 11.0VSSTAANKK363 pKa = 9.92ALQKK367 pKa = 9.44KK368 pKa = 6.15TRR370 pKa = 11.84PTFTIHH376 pKa = 6.79GLRR379 pKa = 11.84HH380 pKa = 5.78TYY382 pKa = 10.53VSFLIYY388 pKa = 10.59NDD390 pKa = 3.95LDD392 pKa = 3.62VVTISKK398 pKa = 10.49LVGHH402 pKa = 7.25KK403 pKa = 9.02DD404 pKa = 2.99TTEE407 pKa = 3.89TLKK410 pKa = 10.27TYY412 pKa = 11.1AHH414 pKa = 6.9LFEE417 pKa = 5.37EE418 pKa = 4.69KK419 pKa = 9.88QAQNFDD425 pKa = 3.9KK426 pKa = 10.72IRR428 pKa = 11.84NLFEE432 pKa = 4.89KK433 pKa = 10.59FGADD437 pKa = 4.09LGQTT441 pKa = 3.65
MM1 pKa = 7.62KK2 pKa = 9.87EE3 pKa = 3.74HH4 pKa = 6.99SIKK7 pKa = 10.6KK8 pKa = 9.92LLTGTITLLSVVTLVACSQSNKK30 pKa = 10.41GIIDD34 pKa = 3.68CRR36 pKa = 11.84IKK38 pKa = 10.68KK39 pKa = 10.47SPTLNFGRR47 pKa = 11.84SEE49 pKa = 3.91RR50 pKa = 11.84KK51 pKa = 9.59ANHH54 pKa = 5.48VWSEE58 pKa = 4.02TCFAVGLFTIPIYY71 pKa = 9.47QKK73 pKa = 10.83RR74 pKa = 11.84KK75 pKa = 7.48GKK77 pKa = 10.54SMAYY81 pKa = 8.96FRR83 pKa = 11.84KK84 pKa = 9.89RR85 pKa = 11.84EE86 pKa = 4.06NGWEE90 pKa = 3.66YY91 pKa = 10.51RR92 pKa = 11.84ISYY95 pKa = 8.24KK96 pKa = 10.93APDD99 pKa = 3.57GSYY102 pKa = 9.91KK103 pKa = 10.36QKK105 pKa = 10.58SKK107 pKa = 11.15SGFRR111 pKa = 11.84TKK113 pKa = 10.73SEE115 pKa = 3.93AVQAASQAEE124 pKa = 4.22IEE126 pKa = 4.31LSSGIVEE133 pKa = 4.78DD134 pKa = 4.43KK135 pKa = 10.91NISLAEE141 pKa = 3.97YY142 pKa = 9.35FEE144 pKa = 4.32KK145 pKa = 10.39WMEE148 pKa = 3.84IHH150 pKa = 6.86KK151 pKa = 10.21KK152 pKa = 9.28PHH154 pKa = 6.29VDD156 pKa = 2.98PEE158 pKa = 4.45TYY160 pKa = 9.81RR161 pKa = 11.84KK162 pKa = 10.35YY163 pKa = 10.95EE164 pKa = 4.03FNLKK168 pKa = 9.81IIKK171 pKa = 9.76EE172 pKa = 4.23YY173 pKa = 9.84YY174 pKa = 10.29QNTKK178 pKa = 10.02LSKK181 pKa = 9.44ITTTMHH187 pKa = 4.98QQVLNKK193 pKa = 10.34LADD196 pKa = 3.98RR197 pKa = 11.84YY198 pKa = 10.75VKK200 pKa = 10.0YY201 pKa = 9.04TVKK204 pKa = 10.61LFNSHH209 pKa = 4.62IRR211 pKa = 11.84AAIKK215 pKa = 10.31VALHH219 pKa = 5.79QGTLKK224 pKa = 10.66KK225 pKa = 10.75DD226 pKa = 3.45FTQLAKK232 pKa = 10.7VFSNVEE238 pKa = 4.17SKK240 pKa = 11.16DD241 pKa = 3.48EE242 pKa = 4.13SEE244 pKa = 6.13KK245 pKa = 10.63YY246 pKa = 10.66LEE248 pKa = 4.3LNGYY252 pKa = 9.96RR253 pKa = 11.84SLLLDD258 pKa = 3.17VRR260 pKa = 11.84QSINHH265 pKa = 5.28QSHH268 pKa = 6.24FFIYY272 pKa = 9.97TIGKK276 pKa = 6.89TGLRR280 pKa = 11.84FSEE283 pKa = 4.07ATGLSKK289 pKa = 10.0FTVDD293 pKa = 3.28RR294 pKa = 11.84QNMCLHH300 pKa = 5.75IRR302 pKa = 11.84KK303 pKa = 7.75TYY305 pKa = 10.1KK306 pKa = 10.6VYY308 pKa = 10.59GKK310 pKa = 10.35KK311 pKa = 10.25KK312 pKa = 9.78GWGPTKK318 pKa = 10.55NPQSEE323 pKa = 3.99RR324 pKa = 11.84DD325 pKa = 3.66VPIDD329 pKa = 3.87RR330 pKa = 11.84DD331 pKa = 3.21WLEE334 pKa = 4.8AYY336 pKa = 10.11DD337 pKa = 4.26EE338 pKa = 4.21YY339 pKa = 11.35MEE341 pKa = 5.0VGYY344 pKa = 10.28IDD346 pKa = 4.5NPEE349 pKa = 3.55KK350 pKa = 10.8RR351 pKa = 11.84LFFKK355 pKa = 11.0VSSTAANKK363 pKa = 9.92ALQKK367 pKa = 9.44KK368 pKa = 6.15TRR370 pKa = 11.84PTFTIHH376 pKa = 6.79GLRR379 pKa = 11.84HH380 pKa = 5.78TYY382 pKa = 10.53VSFLIYY388 pKa = 10.59NDD390 pKa = 3.95LDD392 pKa = 3.62VVTISKK398 pKa = 10.49LVGHH402 pKa = 7.25KK403 pKa = 9.02DD404 pKa = 2.99TTEE407 pKa = 3.89TLKK410 pKa = 10.27TYY412 pKa = 11.1AHH414 pKa = 6.9LFEE417 pKa = 5.37EE418 pKa = 4.69KK419 pKa = 9.88QAQNFDD425 pKa = 3.9KK426 pKa = 10.72IRR428 pKa = 11.84NLFEE432 pKa = 4.89KK433 pKa = 10.59FGADD437 pKa = 4.09LGQTT441 pKa = 3.65
Molecular weight: 51.19 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
10784 |
43 |
1292 |
211.5 |
23.94 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.992 ± 0.508 | 0.566 ± 0.111 |
6.259 ± 0.309 | 7.826 ± 0.555 |
4.238 ± 0.307 | 6.667 ± 0.516 |
1.502 ± 0.177 | 6.714 ± 0.21 |
8.244 ± 0.558 | 7.845 ± 0.41 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.587 ± 0.184 | 5.666 ± 0.29 |
2.68 ± 0.171 | 3.746 ± 0.252 |
4.674 ± 0.239 | 6.417 ± 0.463 |
5.963 ± 0.47 | 6.25 ± 0.314 |
1.345 ± 0.139 | 3.82 ± 0.27 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
