
Beihai sobemo-like virus 20
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 5.58
Get precalculated fractions of proteins
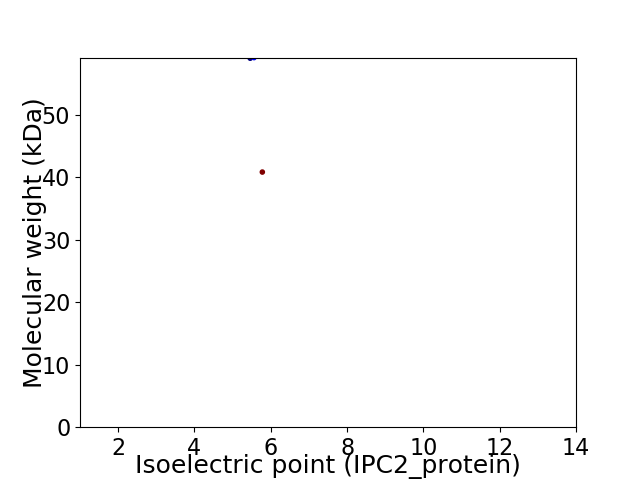
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KE67|A0A1L3KE67_9VIRU Uncharacterized protein OS=Beihai sobemo-like virus 20 OX=1922692 PE=4 SV=1
MM1 pKa = 7.71LDD3 pKa = 3.45SRR5 pKa = 11.84FGVQKK10 pKa = 10.68QEE12 pKa = 4.99SEE14 pKa = 4.23DD15 pKa = 3.74FLNKK19 pKa = 9.68ILEE22 pKa = 4.36SDD24 pKa = 4.0DD25 pKa = 3.63IEE27 pKa = 4.38NVEE30 pKa = 4.92SFVEE34 pKa = 4.2PNFDD38 pKa = 2.94EE39 pKa = 4.78GFRR42 pKa = 11.84VKK44 pKa = 10.45DD45 pKa = 3.55RR46 pKa = 11.84KK47 pKa = 10.7GRR49 pKa = 11.84THH51 pKa = 6.5VFWSDD56 pKa = 3.21NITDD60 pKa = 4.32EE61 pKa = 5.45IVTKK65 pKa = 10.76LNKK68 pKa = 9.53KK69 pKa = 8.81SKK71 pKa = 9.2KK72 pKa = 10.12ASYY75 pKa = 9.57TLTPVQLQSAAQIQPQAADD94 pKa = 3.46NPTPKK99 pKa = 9.39DD100 pKa = 3.39TKK102 pKa = 10.91LSDD105 pKa = 3.5SEE107 pKa = 4.03IQLRR111 pKa = 11.84EE112 pKa = 4.06QINEE116 pKa = 3.84LTKK119 pKa = 10.64EE120 pKa = 4.15VTKK123 pKa = 10.87LRR125 pKa = 11.84EE126 pKa = 3.79EE127 pKa = 4.04QKK129 pKa = 11.11ALNVQPQRR137 pKa = 11.84PVANVDD143 pKa = 3.38YY144 pKa = 10.83QQVQEE149 pKa = 4.5AQVCMGLYY157 pKa = 9.92QRR159 pKa = 11.84ATQILTLAASLQQFLSGHH177 pKa = 5.67FRR179 pKa = 11.84DD180 pKa = 3.87EE181 pKa = 4.01PARR184 pKa = 11.84VKK186 pKa = 10.62QFAANQQLINGVLNQLAQVGYY207 pKa = 11.14AMSAKK212 pKa = 7.74MTTIAQLSDD221 pKa = 3.52EE222 pKa = 4.71DD223 pKa = 3.97LQNKK227 pKa = 7.98NRR229 pKa = 11.84ADD231 pKa = 4.06RR232 pKa = 11.84QRR234 pKa = 11.84KK235 pKa = 8.44KK236 pKa = 10.73IPMPEE241 pKa = 3.8ADD243 pKa = 3.41EE244 pKa = 4.18EE245 pKa = 4.45TRR247 pKa = 11.84NMIEE251 pKa = 4.44EE252 pKa = 4.28ALQFANEE259 pKa = 3.88GFEE262 pKa = 4.35RR263 pKa = 11.84IEE265 pKa = 4.66AIPDD269 pKa = 3.39RR270 pKa = 11.84LYY272 pKa = 10.63PDD274 pKa = 3.81YY275 pKa = 10.77AAPAPPVEE283 pKa = 4.26PQGASDD289 pKa = 4.74YY290 pKa = 10.61ADD292 pKa = 3.84CMAKK296 pKa = 9.97KK297 pKa = 10.47YY298 pKa = 10.48SVQIPTIFQLGSAADD313 pKa = 4.01FLSHH317 pKa = 7.21KK318 pKa = 10.48ASQTQQSEE326 pKa = 4.07GRR328 pKa = 11.84KK329 pKa = 9.54NIRR332 pKa = 11.84TEE334 pKa = 3.94KK335 pKa = 9.03PQMSEE340 pKa = 4.09VEE342 pKa = 4.31KK343 pKa = 10.08QTATLMSNFPGLSKK357 pKa = 10.66KK358 pKa = 10.55LSPTQVATSGPKK370 pKa = 8.35PTEE373 pKa = 3.86PQLPPVSPSTHH384 pKa = 6.35KK385 pKa = 10.87EE386 pKa = 3.75EE387 pKa = 4.45PKK389 pKa = 9.49PAKK392 pKa = 9.28TAWSQQKK399 pKa = 8.97MSEE402 pKa = 4.1PSVQDD407 pKa = 3.05VCANAIEE414 pKa = 4.42MLSATSSQSDD424 pKa = 3.59SLAQALSLLKK434 pKa = 10.62KK435 pKa = 9.44MQLEE439 pKa = 4.14NEE441 pKa = 4.38KK442 pKa = 10.42EE443 pKa = 4.22VEE445 pKa = 4.18SLQEE449 pKa = 3.74EE450 pKa = 4.69GLPRR454 pKa = 11.84SEE456 pKa = 4.34EE457 pKa = 3.89LRR459 pKa = 11.84EE460 pKa = 4.21DD461 pKa = 3.87EE462 pKa = 4.18EE463 pKa = 5.47DD464 pKa = 3.45EE465 pKa = 4.99FILVEE470 pKa = 4.0PQKK473 pKa = 11.26VKK475 pKa = 11.04DD476 pKa = 3.73MKK478 pKa = 10.63KK479 pKa = 10.49CPACHH484 pKa = 6.15SKK486 pKa = 10.45VKK488 pKa = 10.73KK489 pKa = 10.11GDD491 pKa = 2.93TWEE494 pKa = 4.05DD495 pKa = 3.28HH496 pKa = 6.75KK497 pKa = 11.32KK498 pKa = 10.17QPCFVEE504 pKa = 4.51WSKK507 pKa = 11.29KK508 pKa = 9.97RR509 pKa = 11.84PPLNTNKK516 pKa = 9.36VPRR519 pKa = 11.84PKK521 pKa = 8.3TTAA524 pKa = 3.01
MM1 pKa = 7.71LDD3 pKa = 3.45SRR5 pKa = 11.84FGVQKK10 pKa = 10.68QEE12 pKa = 4.99SEE14 pKa = 4.23DD15 pKa = 3.74FLNKK19 pKa = 9.68ILEE22 pKa = 4.36SDD24 pKa = 4.0DD25 pKa = 3.63IEE27 pKa = 4.38NVEE30 pKa = 4.92SFVEE34 pKa = 4.2PNFDD38 pKa = 2.94EE39 pKa = 4.78GFRR42 pKa = 11.84VKK44 pKa = 10.45DD45 pKa = 3.55RR46 pKa = 11.84KK47 pKa = 10.7GRR49 pKa = 11.84THH51 pKa = 6.5VFWSDD56 pKa = 3.21NITDD60 pKa = 4.32EE61 pKa = 5.45IVTKK65 pKa = 10.76LNKK68 pKa = 9.53KK69 pKa = 8.81SKK71 pKa = 9.2KK72 pKa = 10.12ASYY75 pKa = 9.57TLTPVQLQSAAQIQPQAADD94 pKa = 3.46NPTPKK99 pKa = 9.39DD100 pKa = 3.39TKK102 pKa = 10.91LSDD105 pKa = 3.5SEE107 pKa = 4.03IQLRR111 pKa = 11.84EE112 pKa = 4.06QINEE116 pKa = 3.84LTKK119 pKa = 10.64EE120 pKa = 4.15VTKK123 pKa = 10.87LRR125 pKa = 11.84EE126 pKa = 3.79EE127 pKa = 4.04QKK129 pKa = 11.11ALNVQPQRR137 pKa = 11.84PVANVDD143 pKa = 3.38YY144 pKa = 10.83QQVQEE149 pKa = 4.5AQVCMGLYY157 pKa = 9.92QRR159 pKa = 11.84ATQILTLAASLQQFLSGHH177 pKa = 5.67FRR179 pKa = 11.84DD180 pKa = 3.87EE181 pKa = 4.01PARR184 pKa = 11.84VKK186 pKa = 10.62QFAANQQLINGVLNQLAQVGYY207 pKa = 11.14AMSAKK212 pKa = 7.74MTTIAQLSDD221 pKa = 3.52EE222 pKa = 4.71DD223 pKa = 3.97LQNKK227 pKa = 7.98NRR229 pKa = 11.84ADD231 pKa = 4.06RR232 pKa = 11.84QRR234 pKa = 11.84KK235 pKa = 8.44KK236 pKa = 10.73IPMPEE241 pKa = 3.8ADD243 pKa = 3.41EE244 pKa = 4.18EE245 pKa = 4.45TRR247 pKa = 11.84NMIEE251 pKa = 4.44EE252 pKa = 4.28ALQFANEE259 pKa = 3.88GFEE262 pKa = 4.35RR263 pKa = 11.84IEE265 pKa = 4.66AIPDD269 pKa = 3.39RR270 pKa = 11.84LYY272 pKa = 10.63PDD274 pKa = 3.81YY275 pKa = 10.77AAPAPPVEE283 pKa = 4.26PQGASDD289 pKa = 4.74YY290 pKa = 10.61ADD292 pKa = 3.84CMAKK296 pKa = 9.97KK297 pKa = 10.47YY298 pKa = 10.48SVQIPTIFQLGSAADD313 pKa = 4.01FLSHH317 pKa = 7.21KK318 pKa = 10.48ASQTQQSEE326 pKa = 4.07GRR328 pKa = 11.84KK329 pKa = 9.54NIRR332 pKa = 11.84TEE334 pKa = 3.94KK335 pKa = 9.03PQMSEE340 pKa = 4.09VEE342 pKa = 4.31KK343 pKa = 10.08QTATLMSNFPGLSKK357 pKa = 10.66KK358 pKa = 10.55LSPTQVATSGPKK370 pKa = 8.35PTEE373 pKa = 3.86PQLPPVSPSTHH384 pKa = 6.35KK385 pKa = 10.87EE386 pKa = 3.75EE387 pKa = 4.45PKK389 pKa = 9.49PAKK392 pKa = 9.28TAWSQQKK399 pKa = 8.97MSEE402 pKa = 4.1PSVQDD407 pKa = 3.05VCANAIEE414 pKa = 4.42MLSATSSQSDD424 pKa = 3.59SLAQALSLLKK434 pKa = 10.62KK435 pKa = 9.44MQLEE439 pKa = 4.14NEE441 pKa = 4.38KK442 pKa = 10.42EE443 pKa = 4.22VEE445 pKa = 4.18SLQEE449 pKa = 3.74EE450 pKa = 4.69GLPRR454 pKa = 11.84SEE456 pKa = 4.34EE457 pKa = 3.89LRR459 pKa = 11.84EE460 pKa = 4.21DD461 pKa = 3.87EE462 pKa = 4.18EE463 pKa = 5.47DD464 pKa = 3.45EE465 pKa = 4.99FILVEE470 pKa = 4.0PQKK473 pKa = 11.26VKK475 pKa = 11.04DD476 pKa = 3.73MKK478 pKa = 10.63KK479 pKa = 10.49CPACHH484 pKa = 6.15SKK486 pKa = 10.45VKK488 pKa = 10.73KK489 pKa = 10.11GDD491 pKa = 2.93TWEE494 pKa = 4.05DD495 pKa = 3.28HH496 pKa = 6.75KK497 pKa = 11.32KK498 pKa = 10.17QPCFVEE504 pKa = 4.51WSKK507 pKa = 11.29KK508 pKa = 9.97RR509 pKa = 11.84PPLNTNKK516 pKa = 9.36VPRR519 pKa = 11.84PKK521 pKa = 8.3TTAA524 pKa = 3.01
Molecular weight: 59.08 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KE67|A0A1L3KE67_9VIRU Uncharacterized protein OS=Beihai sobemo-like virus 20 OX=1922692 PE=4 SV=1
MM1 pKa = 7.84DD2 pKa = 4.06MVVDD6 pKa = 4.36PTTSPGLPWTSIGTTNAQVFGYY28 pKa = 9.91QEE30 pKa = 4.72GIGFRR35 pKa = 11.84GQQVEE40 pKa = 4.14ILYY43 pKa = 10.55KK44 pKa = 10.47EE45 pKa = 4.27VILRR49 pKa = 11.84LQQLRR54 pKa = 11.84EE55 pKa = 4.15GPKK58 pKa = 9.94MNDD61 pKa = 2.36INLFIKK67 pKa = 10.35PEE69 pKa = 3.69PHH71 pKa = 6.39KK72 pKa = 10.29PKK74 pKa = 10.4KK75 pKa = 10.01VEE77 pKa = 3.62QGAWRR82 pKa = 11.84LIHH85 pKa = 6.31GVSITDD91 pKa = 3.24QMVAEE96 pKa = 4.21IVMGEE101 pKa = 4.29LMDD104 pKa = 5.68LVYY107 pKa = 10.55KK108 pKa = 10.84CPGRR112 pKa = 11.84YY113 pKa = 9.71GPMIGWSPFTTEE125 pKa = 4.89GIPYY129 pKa = 10.17FNFFVGNGPKK139 pKa = 9.89QSADD143 pKa = 3.1KK144 pKa = 11.17SSWDD148 pKa = 3.13WTVQDD153 pKa = 3.36WLAEE157 pKa = 4.06VFIDD161 pKa = 5.15LICYY165 pKa = 9.43IQDD168 pKa = 4.3DD169 pKa = 4.37LNEE172 pKa = 4.33TCSTQLRR179 pKa = 11.84NHH181 pKa = 5.52MRR183 pKa = 11.84AILGRR188 pKa = 11.84KK189 pKa = 8.7VIVGKK194 pKa = 8.01GQRR197 pKa = 11.84FTMDD201 pKa = 2.76GGLPSGWKK209 pKa = 8.47ATIFANSVMQEE220 pKa = 4.27IIHH223 pKa = 6.07HH224 pKa = 4.97VAEE227 pKa = 4.71KK228 pKa = 10.66RR229 pKa = 11.84SGCYY233 pKa = 9.44GKK235 pKa = 9.84PPITMGDD242 pKa = 3.55DD243 pKa = 3.54TLQEE247 pKa = 4.28PMPPAYY253 pKa = 9.27WEE255 pKa = 4.16ALATTGCILKK265 pKa = 10.39EE266 pKa = 4.28VVTHH270 pKa = 6.06QDD272 pKa = 2.6RR273 pKa = 11.84PYY275 pKa = 9.49EE276 pKa = 3.81FCGFHH281 pKa = 6.08FGSRR285 pKa = 11.84EE286 pKa = 3.77YY287 pKa = 10.84EE288 pKa = 3.49PSYY291 pKa = 9.39KK292 pKa = 9.81QKK294 pKa = 10.1HH295 pKa = 5.4AFLVRR300 pKa = 11.84HH301 pKa = 5.92IEE303 pKa = 4.01RR304 pKa = 11.84EE305 pKa = 4.02NFEE308 pKa = 4.23ATIQSYY314 pKa = 10.71QILYY318 pKa = 10.44LFDD321 pKa = 3.62EE322 pKa = 4.69RR323 pKa = 11.84RR324 pKa = 11.84LNALRR329 pKa = 11.84RR330 pKa = 11.84WCEE333 pKa = 3.7LEE335 pKa = 5.05GFAHH339 pKa = 6.62CKK341 pKa = 9.78IDD343 pKa = 3.51VSDD346 pKa = 3.35RR347 pKa = 11.84KK348 pKa = 10.3YY349 pKa = 10.9VVRR352 pKa = 11.84GTPEE356 pKa = 3.18
MM1 pKa = 7.84DD2 pKa = 4.06MVVDD6 pKa = 4.36PTTSPGLPWTSIGTTNAQVFGYY28 pKa = 9.91QEE30 pKa = 4.72GIGFRR35 pKa = 11.84GQQVEE40 pKa = 4.14ILYY43 pKa = 10.55KK44 pKa = 10.47EE45 pKa = 4.27VILRR49 pKa = 11.84LQQLRR54 pKa = 11.84EE55 pKa = 4.15GPKK58 pKa = 9.94MNDD61 pKa = 2.36INLFIKK67 pKa = 10.35PEE69 pKa = 3.69PHH71 pKa = 6.39KK72 pKa = 10.29PKK74 pKa = 10.4KK75 pKa = 10.01VEE77 pKa = 3.62QGAWRR82 pKa = 11.84LIHH85 pKa = 6.31GVSITDD91 pKa = 3.24QMVAEE96 pKa = 4.21IVMGEE101 pKa = 4.29LMDD104 pKa = 5.68LVYY107 pKa = 10.55KK108 pKa = 10.84CPGRR112 pKa = 11.84YY113 pKa = 9.71GPMIGWSPFTTEE125 pKa = 4.89GIPYY129 pKa = 10.17FNFFVGNGPKK139 pKa = 9.89QSADD143 pKa = 3.1KK144 pKa = 11.17SSWDD148 pKa = 3.13WTVQDD153 pKa = 3.36WLAEE157 pKa = 4.06VFIDD161 pKa = 5.15LICYY165 pKa = 9.43IQDD168 pKa = 4.3DD169 pKa = 4.37LNEE172 pKa = 4.33TCSTQLRR179 pKa = 11.84NHH181 pKa = 5.52MRR183 pKa = 11.84AILGRR188 pKa = 11.84KK189 pKa = 8.7VIVGKK194 pKa = 8.01GQRR197 pKa = 11.84FTMDD201 pKa = 2.76GGLPSGWKK209 pKa = 8.47ATIFANSVMQEE220 pKa = 4.27IIHH223 pKa = 6.07HH224 pKa = 4.97VAEE227 pKa = 4.71KK228 pKa = 10.66RR229 pKa = 11.84SGCYY233 pKa = 9.44GKK235 pKa = 9.84PPITMGDD242 pKa = 3.55DD243 pKa = 3.54TLQEE247 pKa = 4.28PMPPAYY253 pKa = 9.27WEE255 pKa = 4.16ALATTGCILKK265 pKa = 10.39EE266 pKa = 4.28VVTHH270 pKa = 6.06QDD272 pKa = 2.6RR273 pKa = 11.84PYY275 pKa = 9.49EE276 pKa = 3.81FCGFHH281 pKa = 6.08FGSRR285 pKa = 11.84EE286 pKa = 3.77YY287 pKa = 10.84EE288 pKa = 3.49PSYY291 pKa = 9.39KK292 pKa = 9.81QKK294 pKa = 10.1HH295 pKa = 5.4AFLVRR300 pKa = 11.84HH301 pKa = 5.92IEE303 pKa = 4.01RR304 pKa = 11.84EE305 pKa = 4.02NFEE308 pKa = 4.23ATIQSYY314 pKa = 10.71QILYY318 pKa = 10.44LFDD321 pKa = 3.62EE322 pKa = 4.69RR323 pKa = 11.84RR324 pKa = 11.84LNALRR329 pKa = 11.84RR330 pKa = 11.84WCEE333 pKa = 3.7LEE335 pKa = 5.05GFAHH339 pKa = 6.62CKK341 pKa = 9.78IDD343 pKa = 3.51VSDD346 pKa = 3.35RR347 pKa = 11.84KK348 pKa = 10.3YY349 pKa = 10.9VVRR352 pKa = 11.84GTPEE356 pKa = 3.18
Molecular weight: 40.86 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
880 |
356 |
524 |
440.0 |
49.97 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.705 ± 1.287 | 1.591 ± 0.382 |
5.341 ± 0.166 | 8.636 ± 0.776 |
3.75 ± 0.597 | 5.114 ± 1.929 |
1.818 ± 0.577 | 5.0 ± 1.177 |
7.614 ± 1.325 | 7.273 ± 0.309 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.841 ± 0.308 | 3.636 ± 0.482 |
6.591 ± 0.403 | 7.614 ± 1.325 |
4.773 ± 0.492 | 6.25 ± 1.185 |
5.568 ± 0.029 | 5.909 ± 0.321 |
1.477 ± 0.612 | 2.5 ± 0.834 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
