
Shahe tombus-like virus 1
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.87
Get precalculated fractions of proteins
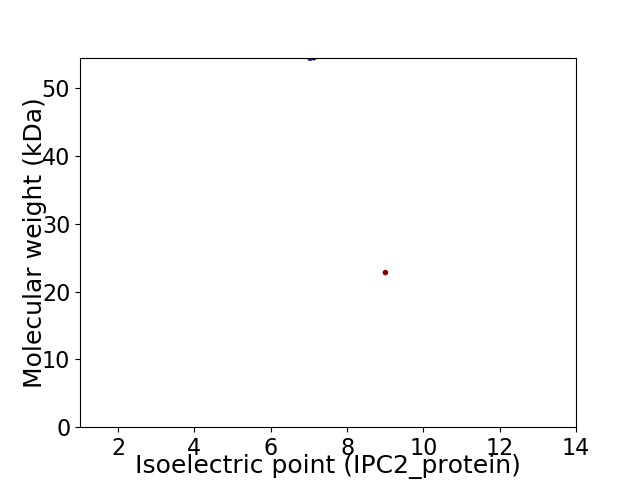
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KGH4|A0A1L3KGH4_9VIRU RNA-directed RNA polymerase OS=Shahe tombus-like virus 1 OX=1923455 PE=4 SV=1
MM1 pKa = 8.04RR2 pKa = 11.84YY3 pKa = 9.28LAGFGTGVRR12 pKa = 11.84YY13 pKa = 9.22GVHH16 pKa = 6.16CFNYY20 pKa = 9.13TNLARR25 pKa = 11.84GIVEE29 pKa = 3.66RR30 pKa = 11.84VFYY33 pKa = 10.51VRR35 pKa = 11.84RR36 pKa = 11.84GEE38 pKa = 4.3GLAPAPQPSVGVFSRR53 pKa = 11.84LNAVRR58 pKa = 11.84QRR60 pKa = 11.84LLRR63 pKa = 11.84AVRR66 pKa = 11.84PTPVVARR73 pKa = 11.84DD74 pKa = 4.12EE75 pKa = 4.45YY76 pKa = 10.37PGLYY80 pKa = 9.16TGRR83 pKa = 11.84KK84 pKa = 6.95RR85 pKa = 11.84GVYY88 pKa = 9.29EE89 pKa = 4.09RR90 pKa = 11.84ALEE93 pKa = 4.1SLKK96 pKa = 10.48VRR98 pKa = 11.84AINFRR103 pKa = 11.84DD104 pKa = 2.98AWVNTFVKK112 pKa = 10.53AEE114 pKa = 4.02KK115 pKa = 10.82VNLDD119 pKa = 3.82SKK121 pKa = 11.25GDD123 pKa = 3.66PAPRR127 pKa = 11.84VIQPRR132 pKa = 11.84SPRR135 pKa = 11.84YY136 pKa = 7.87NLEE139 pKa = 3.41VGRR142 pKa = 11.84YY143 pKa = 8.32LKK145 pKa = 10.84LFEE148 pKa = 5.39KK149 pKa = 9.88EE150 pKa = 3.56LCAGFEE156 pKa = 4.39RR157 pKa = 11.84VWGYY161 pKa = 9.91PVVLKK166 pKa = 10.78GKK168 pKa = 9.18NAQDD172 pKa = 3.14VGAAMADD179 pKa = 3.18HH180 pKa = 7.15WQQFAQPVAVGLDD193 pKa = 3.16ASRR196 pKa = 11.84FDD198 pKa = 3.43QHH200 pKa = 6.33VSKK203 pKa = 11.0DD204 pKa = 3.37ALRR207 pKa = 11.84WEE209 pKa = 4.13HH210 pKa = 6.33SVYY213 pKa = 10.99NSVFNSPEE221 pKa = 3.92LRR223 pKa = 11.84RR224 pKa = 11.84LLQWQLRR231 pKa = 11.84NHH233 pKa = 7.01GIARR237 pKa = 11.84VEE239 pKa = 4.02DD240 pKa = 4.69LIVEE244 pKa = 4.0YY245 pKa = 10.41DD246 pKa = 3.32IEE248 pKa = 4.54GCRR251 pKa = 11.84MSGDD255 pKa = 3.38INTGLGNCLIMSSIVIAYY273 pKa = 9.24CEE275 pKa = 3.5QAGIYY280 pKa = 9.75FRR282 pKa = 11.84LANNGDD288 pKa = 3.76DD289 pKa = 4.07CVLFVDD295 pKa = 5.11KK296 pKa = 11.27ADD298 pKa = 4.1LGALAGIDD306 pKa = 3.32QWFLDD311 pKa = 3.99FGFTLTRR318 pKa = 11.84EE319 pKa = 4.02EE320 pKa = 3.98PVYY323 pKa = 10.98VLEE326 pKa = 4.87QVVFCQAQPVLTSTGWRR343 pKa = 11.84MVRR346 pKa = 11.84DD347 pKa = 3.68PRR349 pKa = 11.84TAMSKK354 pKa = 10.71DD355 pKa = 3.63CVSLLGWDD363 pKa = 3.53NAQSFKK369 pKa = 10.83LWASAIGACGLSLTRR384 pKa = 11.84GVPVWEE390 pKa = 3.92AWYY393 pKa = 10.75SRR395 pKa = 11.84LVHH398 pKa = 6.59LGAGAVSQGVADD410 pKa = 4.42HH411 pKa = 6.43VWDD414 pKa = 3.87SGLGYY419 pKa = 9.67MSRR422 pKa = 11.84GVLGGEE428 pKa = 3.78VDD430 pKa = 3.31AEE432 pKa = 4.2ARR434 pKa = 11.84VSFYY438 pKa = 10.7RR439 pKa = 11.84AFGILPDD446 pKa = 3.75LQEE449 pKa = 4.3ALEE452 pKa = 4.48AEE454 pKa = 5.06YY455 pKa = 10.71ACPMPIGSPTPMMHH469 pKa = 6.79PSIVAIDD476 pKa = 3.82TTCNPLATWLAAKK489 pKa = 10.11
MM1 pKa = 8.04RR2 pKa = 11.84YY3 pKa = 9.28LAGFGTGVRR12 pKa = 11.84YY13 pKa = 9.22GVHH16 pKa = 6.16CFNYY20 pKa = 9.13TNLARR25 pKa = 11.84GIVEE29 pKa = 3.66RR30 pKa = 11.84VFYY33 pKa = 10.51VRR35 pKa = 11.84RR36 pKa = 11.84GEE38 pKa = 4.3GLAPAPQPSVGVFSRR53 pKa = 11.84LNAVRR58 pKa = 11.84QRR60 pKa = 11.84LLRR63 pKa = 11.84AVRR66 pKa = 11.84PTPVVARR73 pKa = 11.84DD74 pKa = 4.12EE75 pKa = 4.45YY76 pKa = 10.37PGLYY80 pKa = 9.16TGRR83 pKa = 11.84KK84 pKa = 6.95RR85 pKa = 11.84GVYY88 pKa = 9.29EE89 pKa = 4.09RR90 pKa = 11.84ALEE93 pKa = 4.1SLKK96 pKa = 10.48VRR98 pKa = 11.84AINFRR103 pKa = 11.84DD104 pKa = 2.98AWVNTFVKK112 pKa = 10.53AEE114 pKa = 4.02KK115 pKa = 10.82VNLDD119 pKa = 3.82SKK121 pKa = 11.25GDD123 pKa = 3.66PAPRR127 pKa = 11.84VIQPRR132 pKa = 11.84SPRR135 pKa = 11.84YY136 pKa = 7.87NLEE139 pKa = 3.41VGRR142 pKa = 11.84YY143 pKa = 8.32LKK145 pKa = 10.84LFEE148 pKa = 5.39KK149 pKa = 9.88EE150 pKa = 3.56LCAGFEE156 pKa = 4.39RR157 pKa = 11.84VWGYY161 pKa = 9.91PVVLKK166 pKa = 10.78GKK168 pKa = 9.18NAQDD172 pKa = 3.14VGAAMADD179 pKa = 3.18HH180 pKa = 7.15WQQFAQPVAVGLDD193 pKa = 3.16ASRR196 pKa = 11.84FDD198 pKa = 3.43QHH200 pKa = 6.33VSKK203 pKa = 11.0DD204 pKa = 3.37ALRR207 pKa = 11.84WEE209 pKa = 4.13HH210 pKa = 6.33SVYY213 pKa = 10.99NSVFNSPEE221 pKa = 3.92LRR223 pKa = 11.84RR224 pKa = 11.84LLQWQLRR231 pKa = 11.84NHH233 pKa = 7.01GIARR237 pKa = 11.84VEE239 pKa = 4.02DD240 pKa = 4.69LIVEE244 pKa = 4.0YY245 pKa = 10.41DD246 pKa = 3.32IEE248 pKa = 4.54GCRR251 pKa = 11.84MSGDD255 pKa = 3.38INTGLGNCLIMSSIVIAYY273 pKa = 9.24CEE275 pKa = 3.5QAGIYY280 pKa = 9.75FRR282 pKa = 11.84LANNGDD288 pKa = 3.76DD289 pKa = 4.07CVLFVDD295 pKa = 5.11KK296 pKa = 11.27ADD298 pKa = 4.1LGALAGIDD306 pKa = 3.32QWFLDD311 pKa = 3.99FGFTLTRR318 pKa = 11.84EE319 pKa = 4.02EE320 pKa = 3.98PVYY323 pKa = 10.98VLEE326 pKa = 4.87QVVFCQAQPVLTSTGWRR343 pKa = 11.84MVRR346 pKa = 11.84DD347 pKa = 3.68PRR349 pKa = 11.84TAMSKK354 pKa = 10.71DD355 pKa = 3.63CVSLLGWDD363 pKa = 3.53NAQSFKK369 pKa = 10.83LWASAIGACGLSLTRR384 pKa = 11.84GVPVWEE390 pKa = 3.92AWYY393 pKa = 10.75SRR395 pKa = 11.84LVHH398 pKa = 6.59LGAGAVSQGVADD410 pKa = 4.42HH411 pKa = 6.43VWDD414 pKa = 3.87SGLGYY419 pKa = 9.67MSRR422 pKa = 11.84GVLGGEE428 pKa = 3.78VDD430 pKa = 3.31AEE432 pKa = 4.2ARR434 pKa = 11.84VSFYY438 pKa = 10.7RR439 pKa = 11.84AFGILPDD446 pKa = 3.75LQEE449 pKa = 4.3ALEE452 pKa = 4.48AEE454 pKa = 5.06YY455 pKa = 10.71ACPMPIGSPTPMMHH469 pKa = 6.79PSIVAIDD476 pKa = 3.82TTCNPLATWLAAKK489 pKa = 10.11
Molecular weight: 54.44 kDa
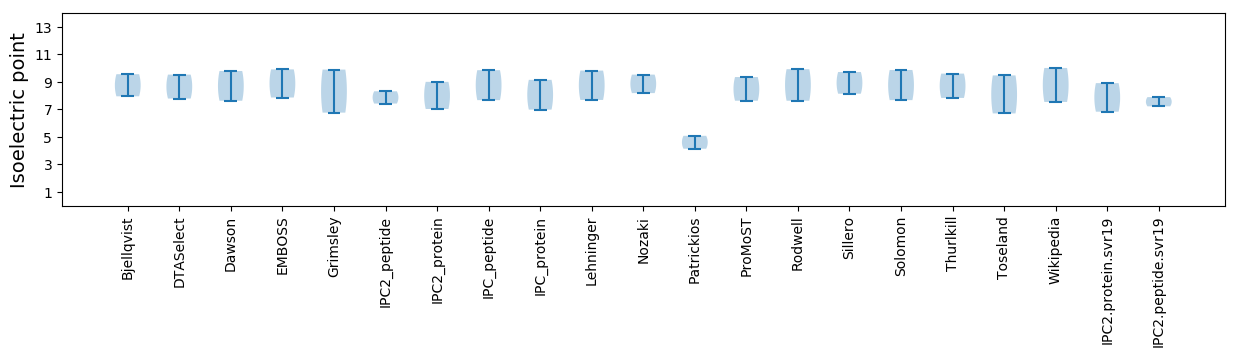
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KGH4|A0A1L3KGH4_9VIRU RNA-directed RNA polymerase OS=Shahe tombus-like virus 1 OX=1923455 PE=4 SV=1
MM1 pKa = 7.65ARR3 pKa = 11.84RR4 pKa = 11.84KK5 pKa = 10.0VMPSPVQGGVRR16 pKa = 11.84RR17 pKa = 11.84RR18 pKa = 11.84RR19 pKa = 11.84NKK21 pKa = 9.79QPNVTQTSGTSSVIKK36 pKa = 10.28YY37 pKa = 9.36SALGSTVTSDD47 pKa = 3.5VNGEE51 pKa = 3.59AWYY54 pKa = 9.86HH55 pKa = 5.53RR56 pKa = 11.84MYY58 pKa = 10.77IPGSFGGLTQTVGTAIVGYY77 pKa = 10.04YY78 pKa = 8.85STAKK82 pKa = 9.82FQAGTTLRR90 pKa = 11.84WEE92 pKa = 4.26PSVSFSTSGRR102 pKa = 11.84VYY104 pKa = 11.03VGFTDD109 pKa = 3.53NPEE112 pKa = 4.1MIEE115 pKa = 3.91AMRR118 pKa = 11.84VLTGGNAVNAVKK130 pKa = 10.77GLGDD134 pKa = 3.86VISFPVWQEE143 pKa = 3.06TDD145 pKa = 3.01IPFSPRR151 pKa = 11.84MRR153 pKa = 11.84RR154 pKa = 11.84KK155 pKa = 9.4MFDD158 pKa = 3.15TNQAAITGVDD168 pKa = 3.53VLDD171 pKa = 4.3RR172 pKa = 11.84TCQTAMFLAIDD183 pKa = 4.33GMPASTRR190 pKa = 11.84AGSVWFHH197 pKa = 7.27DD198 pKa = 3.9NVAVEE203 pKa = 4.42GMSSIEE209 pKa = 4.03TT210 pKa = 3.67
MM1 pKa = 7.65ARR3 pKa = 11.84RR4 pKa = 11.84KK5 pKa = 10.0VMPSPVQGGVRR16 pKa = 11.84RR17 pKa = 11.84RR18 pKa = 11.84RR19 pKa = 11.84NKK21 pKa = 9.79QPNVTQTSGTSSVIKK36 pKa = 10.28YY37 pKa = 9.36SALGSTVTSDD47 pKa = 3.5VNGEE51 pKa = 3.59AWYY54 pKa = 9.86HH55 pKa = 5.53RR56 pKa = 11.84MYY58 pKa = 10.77IPGSFGGLTQTVGTAIVGYY77 pKa = 10.04YY78 pKa = 8.85STAKK82 pKa = 9.82FQAGTTLRR90 pKa = 11.84WEE92 pKa = 4.26PSVSFSTSGRR102 pKa = 11.84VYY104 pKa = 11.03VGFTDD109 pKa = 3.53NPEE112 pKa = 4.1MIEE115 pKa = 3.91AMRR118 pKa = 11.84VLTGGNAVNAVKK130 pKa = 10.77GLGDD134 pKa = 3.86VISFPVWQEE143 pKa = 3.06TDD145 pKa = 3.01IPFSPRR151 pKa = 11.84MRR153 pKa = 11.84RR154 pKa = 11.84KK155 pKa = 9.4MFDD158 pKa = 3.15TNQAAITGVDD168 pKa = 3.53VLDD171 pKa = 4.3RR172 pKa = 11.84TCQTAMFLAIDD183 pKa = 4.33GMPASTRR190 pKa = 11.84AGSVWFHH197 pKa = 7.27DD198 pKa = 3.9NVAVEE203 pKa = 4.42GMSSIEE209 pKa = 4.03TT210 pKa = 3.67
Molecular weight: 22.82 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
699 |
210 |
489 |
349.5 |
38.63 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.156 ± 0.855 | 1.717 ± 0.69 |
5.007 ± 0.401 | 4.578 ± 0.692 |
4.149 ± 0.076 | 9.156 ± 0.469 |
1.431 ± 0.266 | 3.72 ± 0.315 |
2.861 ± 0.002 | 7.582 ± 2.363 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.861 ± 1.057 | 3.577 ± 0.13 |
4.864 ± 0.057 | 3.72 ± 0.05 |
7.725 ± 0.324 | 6.438 ± 1.451 |
5.293 ± 2.618 | 10.157 ± 0.177 |
2.432 ± 0.293 | 3.577 ± 0.4 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
