
Streptomyces phage Daubenski
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Siphoviridae; Samistivirus; unclassified Samistivirus
Average proteome isoelectric point is 6.34
Get precalculated fractions of proteins
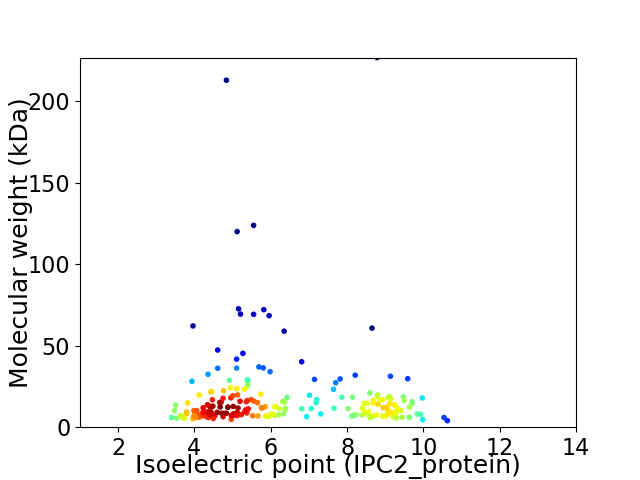
Virtual 2D-PAGE plot for 207 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A5Q2WHL3|A0A5Q2WHL3_9CAUD Uncharacterized protein OS=Streptomyces phage Daubenski OX=2653725 GN=63 PE=4 SV=1
MM1 pKa = 7.76SYY3 pKa = 10.54QLQVLADD10 pKa = 3.61NPFSYY15 pKa = 9.85WKK17 pKa = 10.48LDD19 pKa = 3.37EE20 pKa = 4.85TGPAFPDD27 pKa = 3.31SAGSLRR33 pKa = 11.84TADD36 pKa = 3.61LVGTITRR43 pKa = 11.84HH44 pKa = 5.3PALVTGSGSALLLNNTNQLQMDD66 pKa = 4.02DD67 pKa = 3.63PVFNKK72 pKa = 10.34GYY74 pKa = 8.92EE75 pKa = 3.89LRR77 pKa = 11.84QFSLEE82 pKa = 3.89AWVKK86 pKa = 8.64PVNVTGEE93 pKa = 4.22VALMSHH99 pKa = 6.44SSTYY103 pKa = 10.64DD104 pKa = 3.18GLVISPTNITFRR116 pKa = 11.84TKK118 pKa = 10.49YY119 pKa = 8.85LTAGTCEE126 pKa = 3.71VTYY129 pKa = 10.18EE130 pKa = 3.94YY131 pKa = 10.85DD132 pKa = 3.12ASKK135 pKa = 10.6SFHH138 pKa = 6.0VVGVHH143 pKa = 5.58TNAKK147 pKa = 9.52NSLYY151 pKa = 10.73VDD153 pKa = 4.01GVLVAEE159 pKa = 4.55TDD161 pKa = 3.61LTDD164 pKa = 3.53EE165 pKa = 4.29QQTDD169 pKa = 3.53VYY171 pKa = 11.61AFLTTDD177 pKa = 4.78LIAGQSSGVSTIALDD192 pKa = 3.65APAIYY197 pKa = 9.7TFALTRR203 pKa = 11.84DD204 pKa = 3.93AVAKK208 pKa = 10.25HH209 pKa = 5.74YY210 pKa = 11.12GFGIDD215 pKa = 3.23VDD217 pKa = 4.19SSEE220 pKa = 5.43GIAGYY225 pKa = 10.83NDD227 pKa = 4.38AIFWNFSDD235 pKa = 3.5EE236 pKa = 4.23TRR238 pKa = 11.84NIALKK243 pKa = 9.71QLWSSAADD251 pKa = 3.34WATGTLSDD259 pKa = 4.4TVIIDD264 pKa = 3.41DD265 pKa = 5.04TIAPGYY271 pKa = 8.47SQSEE275 pKa = 4.41SSVVEE280 pKa = 4.34DD281 pKa = 3.83GLIIPVYY288 pKa = 10.11TNTSLPGTWQASIAFNVIPEE308 pKa = 4.35TTISDD313 pKa = 4.26ARR315 pKa = 11.84INWKK319 pKa = 9.71GQGSYY324 pKa = 9.76TIEE327 pKa = 3.92YY328 pKa = 9.37SLDD331 pKa = 3.53GGTVWLTPSDD341 pKa = 3.83SGATTLDD348 pKa = 3.29NDD350 pKa = 3.64STIDD354 pKa = 3.59IRR356 pKa = 11.84ITFDD360 pKa = 2.97GGIVDD365 pKa = 3.79DD366 pKa = 4.41TSFVEE371 pKa = 4.57YY372 pKa = 10.33IEE374 pKa = 4.29VVAYY378 pKa = 10.01LDD380 pKa = 3.38KK381 pKa = 10.99YY382 pKa = 10.84FRR384 pKa = 11.84GSRR387 pKa = 11.84SDD389 pKa = 2.93RR390 pKa = 11.84TAQMSGTGVASDD402 pKa = 3.94NYY404 pKa = 10.54YY405 pKa = 10.98EE406 pKa = 4.7PIEE409 pKa = 4.06YY410 pKa = 10.75ADD412 pKa = 3.5INGLRR417 pKa = 11.84MISGAINVSLDD428 pKa = 3.04SSYY431 pKa = 11.6AGEE434 pKa = 4.52EE435 pKa = 4.18EE436 pKa = 4.57PGDD439 pKa = 3.61LFIRR443 pKa = 11.84GFDD446 pKa = 3.31MWVKK450 pKa = 8.58PTAGNILNATGVNVEE465 pKa = 4.15RR466 pKa = 11.84SGNTIVFSGFSAFTVNGASVSSGATVFGSDD496 pKa = 2.61SWYY499 pKa = 10.47HH500 pKa = 4.92IAGVFTIPANLALQIGSPGIIIAQLSAIYY529 pKa = 8.38DD530 pKa = 4.18TINLAGLQQMYY541 pKa = 10.23AAYY544 pKa = 10.26LGLPGLVVNDD554 pKa = 4.13SSDD557 pKa = 3.48VGIDD561 pKa = 3.2EE562 pKa = 4.72ATPTTNLYY570 pKa = 9.67AHH572 pKa = 6.59VWGITPAGG580 pKa = 3.48
MM1 pKa = 7.76SYY3 pKa = 10.54QLQVLADD10 pKa = 3.61NPFSYY15 pKa = 9.85WKK17 pKa = 10.48LDD19 pKa = 3.37EE20 pKa = 4.85TGPAFPDD27 pKa = 3.31SAGSLRR33 pKa = 11.84TADD36 pKa = 3.61LVGTITRR43 pKa = 11.84HH44 pKa = 5.3PALVTGSGSALLLNNTNQLQMDD66 pKa = 4.02DD67 pKa = 3.63PVFNKK72 pKa = 10.34GYY74 pKa = 8.92EE75 pKa = 3.89LRR77 pKa = 11.84QFSLEE82 pKa = 3.89AWVKK86 pKa = 8.64PVNVTGEE93 pKa = 4.22VALMSHH99 pKa = 6.44SSTYY103 pKa = 10.64DD104 pKa = 3.18GLVISPTNITFRR116 pKa = 11.84TKK118 pKa = 10.49YY119 pKa = 8.85LTAGTCEE126 pKa = 3.71VTYY129 pKa = 10.18EE130 pKa = 3.94YY131 pKa = 10.85DD132 pKa = 3.12ASKK135 pKa = 10.6SFHH138 pKa = 6.0VVGVHH143 pKa = 5.58TNAKK147 pKa = 9.52NSLYY151 pKa = 10.73VDD153 pKa = 4.01GVLVAEE159 pKa = 4.55TDD161 pKa = 3.61LTDD164 pKa = 3.53EE165 pKa = 4.29QQTDD169 pKa = 3.53VYY171 pKa = 11.61AFLTTDD177 pKa = 4.78LIAGQSSGVSTIALDD192 pKa = 3.65APAIYY197 pKa = 9.7TFALTRR203 pKa = 11.84DD204 pKa = 3.93AVAKK208 pKa = 10.25HH209 pKa = 5.74YY210 pKa = 11.12GFGIDD215 pKa = 3.23VDD217 pKa = 4.19SSEE220 pKa = 5.43GIAGYY225 pKa = 10.83NDD227 pKa = 4.38AIFWNFSDD235 pKa = 3.5EE236 pKa = 4.23TRR238 pKa = 11.84NIALKK243 pKa = 9.71QLWSSAADD251 pKa = 3.34WATGTLSDD259 pKa = 4.4TVIIDD264 pKa = 3.41DD265 pKa = 5.04TIAPGYY271 pKa = 8.47SQSEE275 pKa = 4.41SSVVEE280 pKa = 4.34DD281 pKa = 3.83GLIIPVYY288 pKa = 10.11TNTSLPGTWQASIAFNVIPEE308 pKa = 4.35TTISDD313 pKa = 4.26ARR315 pKa = 11.84INWKK319 pKa = 9.71GQGSYY324 pKa = 9.76TIEE327 pKa = 3.92YY328 pKa = 9.37SLDD331 pKa = 3.53GGTVWLTPSDD341 pKa = 3.83SGATTLDD348 pKa = 3.29NDD350 pKa = 3.64STIDD354 pKa = 3.59IRR356 pKa = 11.84ITFDD360 pKa = 2.97GGIVDD365 pKa = 3.79DD366 pKa = 4.41TSFVEE371 pKa = 4.57YY372 pKa = 10.33IEE374 pKa = 4.29VVAYY378 pKa = 10.01LDD380 pKa = 3.38KK381 pKa = 10.99YY382 pKa = 10.84FRR384 pKa = 11.84GSRR387 pKa = 11.84SDD389 pKa = 2.93RR390 pKa = 11.84TAQMSGTGVASDD402 pKa = 3.94NYY404 pKa = 10.54YY405 pKa = 10.98EE406 pKa = 4.7PIEE409 pKa = 4.06YY410 pKa = 10.75ADD412 pKa = 3.5INGLRR417 pKa = 11.84MISGAINVSLDD428 pKa = 3.04SSYY431 pKa = 11.6AGEE434 pKa = 4.52EE435 pKa = 4.18EE436 pKa = 4.57PGDD439 pKa = 3.61LFIRR443 pKa = 11.84GFDD446 pKa = 3.31MWVKK450 pKa = 8.58PTAGNILNATGVNVEE465 pKa = 4.15RR466 pKa = 11.84SGNTIVFSGFSAFTVNGASVSSGATVFGSDD496 pKa = 2.61SWYY499 pKa = 10.47HH500 pKa = 4.92IAGVFTIPANLALQIGSPGIIIAQLSAIYY529 pKa = 8.38DD530 pKa = 4.18TINLAGLQQMYY541 pKa = 10.23AAYY544 pKa = 10.26LGLPGLVVNDD554 pKa = 4.13SSDD557 pKa = 3.48VGIDD561 pKa = 3.2EE562 pKa = 4.72ATPTTNLYY570 pKa = 9.67AHH572 pKa = 6.59VWGITPAGG580 pKa = 3.48
Molecular weight: 62.18 kDa
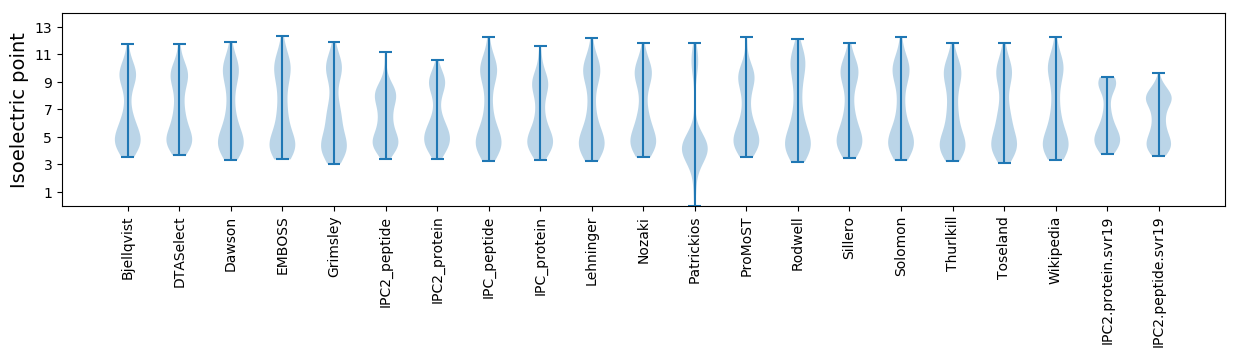
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5Q2WIB1|A0A5Q2WIB1_9CAUD Uncharacterized protein OS=Streptomyces phage Daubenski OX=2653725 GN=108 PE=4 SV=1
MM1 pKa = 7.74ALTGTNGQCWFQRR14 pKa = 11.84CSRR17 pKa = 11.84KK18 pKa = 8.66ATTTIRR24 pKa = 11.84YY25 pKa = 7.5YY26 pKa = 10.01NKK28 pKa = 10.43SVVVCNGHH36 pKa = 6.55KK37 pKa = 10.75NLDD40 pKa = 3.8GVGCPSSRR48 pKa = 11.84RR49 pKa = 11.84RR50 pKa = 11.84PKK52 pKa = 10.46SS53 pKa = 3.11
MM1 pKa = 7.74ALTGTNGQCWFQRR14 pKa = 11.84CSRR17 pKa = 11.84KK18 pKa = 8.66ATTTIRR24 pKa = 11.84YY25 pKa = 7.5YY26 pKa = 10.01NKK28 pKa = 10.43SVVVCNGHH36 pKa = 6.55KK37 pKa = 10.75NLDD40 pKa = 3.8GVGCPSSRR48 pKa = 11.84RR49 pKa = 11.84RR50 pKa = 11.84PKK52 pKa = 10.46SS53 pKa = 3.11
Molecular weight: 5.93 kDa
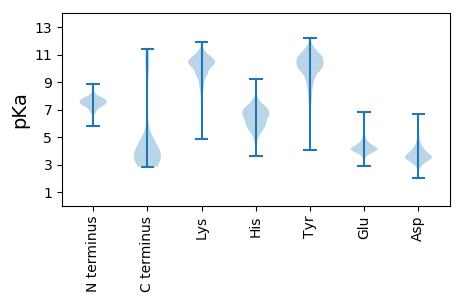
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
35038 |
33 |
2097 |
169.3 |
18.98 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.491 ± 0.352 | 1.053 ± 0.124 |
6.596 ± 0.187 | 6.756 ± 0.315 |
3.819 ± 0.124 | 7.435 ± 0.197 |
1.918 ± 0.155 | 5.271 ± 0.138 |
6.071 ± 0.314 | 7.301 ± 0.197 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.905 ± 0.126 | 4.532 ± 0.141 |
3.722 ± 0.161 | 3.228 ± 0.18 |
5.702 ± 0.198 | 6.016 ± 0.277 |
6.316 ± 0.349 | 7.226 ± 0.216 |
1.889 ± 0.097 | 3.753 ± 0.176 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
