
Thioclava indica
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Rhodobacteraceae; Thioclava
Average proteome isoelectric point is 6.42
Get precalculated fractions of proteins
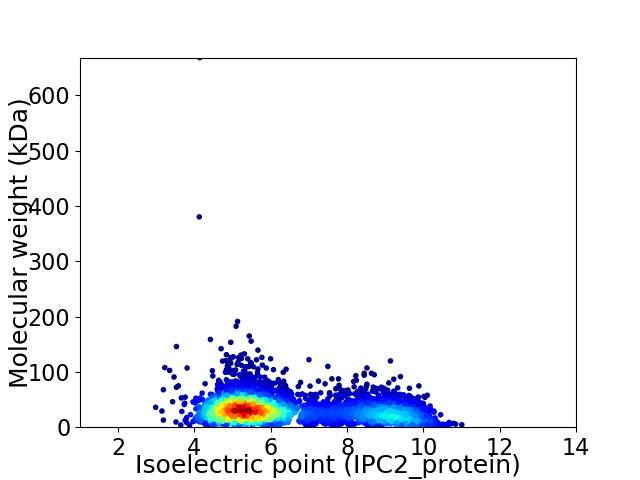
Virtual 2D-PAGE plot for 3671 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A074JT76|A0A074JT76_9RHOB Uncharacterized protein OS=Thioclava indica OX=1353528 GN=DT23_11805 PE=4 SV=1
MM1 pKa = 6.62TVYY4 pKa = 10.76YY5 pKa = 10.83VDD7 pKa = 3.71TNGNDD12 pKa = 3.32NANGSDD18 pKa = 3.52GSPFRR23 pKa = 11.84TIGQALNANLNPGDD37 pKa = 4.27EE38 pKa = 4.31IVVRR42 pKa = 11.84AGTYY46 pKa = 10.15NEE48 pKa = 4.56SLNINQGGSAAGDD61 pKa = 3.19ITIRR65 pKa = 11.84AEE67 pKa = 4.27DD68 pKa = 4.39PGTALIRR75 pKa = 11.84PPSGAWNAISVNANYY90 pKa = 8.52VTIEE94 pKa = 4.05GFDD97 pKa = 3.4IANAAGDD104 pKa = 4.22GIEE107 pKa = 4.42ANSVHH112 pKa = 7.04HH113 pKa = 6.05ITIADD118 pKa = 3.7NTVHH122 pKa = 6.71GNGEE126 pKa = 3.92SGIQFNYY133 pKa = 10.8SEE135 pKa = 5.39FITIEE140 pKa = 3.94GNEE143 pKa = 4.34TYY145 pKa = 11.16DD146 pKa = 3.52NASSGWYY153 pKa = 9.62SGISIYY159 pKa = 8.08QNRR162 pKa = 11.84NVSGDD167 pKa = 3.38TTTPGYY173 pKa = 9.03RR174 pKa = 11.84TIIRR178 pKa = 11.84DD179 pKa = 3.93NITHH183 pKa = 7.32DD184 pKa = 3.46NVTQSGAHH192 pKa = 5.79TDD194 pKa = 3.48GNGIIIDD201 pKa = 4.63DD202 pKa = 4.21FQSTQTDD209 pKa = 3.02GHH211 pKa = 6.63PNYY214 pKa = 9.89TYY216 pKa = 7.8PTLVEE221 pKa = 4.09NNVAYY226 pKa = 10.49GNGGKK231 pKa = 10.25GIQVTWSDD239 pKa = 4.09YY240 pKa = 8.98VTVSNNTAYY249 pKa = 10.6HH250 pKa = 6.09NNVDD254 pKa = 4.17LQNDD258 pKa = 4.25GTWRR262 pKa = 11.84GEE264 pKa = 3.59ISNAQSSNNTFVNNIAVADD283 pKa = 4.02PSINSNNTAIDD294 pKa = 3.52NNSYY298 pKa = 11.01GGYY301 pKa = 10.41SNDD304 pKa = 3.04NVVWEE309 pKa = 4.96DD310 pKa = 3.18NVLYY314 pKa = 10.54NGKK317 pKa = 7.58TGQMSASTDD326 pKa = 3.34GGNDD330 pKa = 3.22APSAADD336 pKa = 3.49GNLIGVNPGFVNPANGDD353 pKa = 3.53FSISAGSVAVNLGADD368 pKa = 3.62MDD370 pKa = 4.49GSSSVTSPPTTSTPDD385 pKa = 3.47PVVTPDD391 pKa = 4.04PDD393 pKa = 3.8PVVTPDD399 pKa = 3.97PAPTTPAVEE408 pKa = 3.99NAAPVAHH415 pKa = 7.47DD416 pKa = 5.23DD417 pKa = 4.05INFKK421 pKa = 10.09TSPNAPLILQQSQLLSNDD439 pKa = 2.81SDD441 pKa = 4.33ADD443 pKa = 3.48GDD445 pKa = 4.52ALILTNVVGSTQGSAKK461 pKa = 9.16LTSGNNISFTPEE473 pKa = 3.17SDD475 pKa = 3.36ASGTASVTYY484 pKa = 9.4TVSDD488 pKa = 3.66QNGGTDD494 pKa = 3.03TATAYY499 pKa = 9.98IDD501 pKa = 3.47IVAAAPSGPGGG512 pKa = 3.39
MM1 pKa = 6.62TVYY4 pKa = 10.76YY5 pKa = 10.83VDD7 pKa = 3.71TNGNDD12 pKa = 3.32NANGSDD18 pKa = 3.52GSPFRR23 pKa = 11.84TIGQALNANLNPGDD37 pKa = 4.27EE38 pKa = 4.31IVVRR42 pKa = 11.84AGTYY46 pKa = 10.15NEE48 pKa = 4.56SLNINQGGSAAGDD61 pKa = 3.19ITIRR65 pKa = 11.84AEE67 pKa = 4.27DD68 pKa = 4.39PGTALIRR75 pKa = 11.84PPSGAWNAISVNANYY90 pKa = 8.52VTIEE94 pKa = 4.05GFDD97 pKa = 3.4IANAAGDD104 pKa = 4.22GIEE107 pKa = 4.42ANSVHH112 pKa = 7.04HH113 pKa = 6.05ITIADD118 pKa = 3.7NTVHH122 pKa = 6.71GNGEE126 pKa = 3.92SGIQFNYY133 pKa = 10.8SEE135 pKa = 5.39FITIEE140 pKa = 3.94GNEE143 pKa = 4.34TYY145 pKa = 11.16DD146 pKa = 3.52NASSGWYY153 pKa = 9.62SGISIYY159 pKa = 8.08QNRR162 pKa = 11.84NVSGDD167 pKa = 3.38TTTPGYY173 pKa = 9.03RR174 pKa = 11.84TIIRR178 pKa = 11.84DD179 pKa = 3.93NITHH183 pKa = 7.32DD184 pKa = 3.46NVTQSGAHH192 pKa = 5.79TDD194 pKa = 3.48GNGIIIDD201 pKa = 4.63DD202 pKa = 4.21FQSTQTDD209 pKa = 3.02GHH211 pKa = 6.63PNYY214 pKa = 9.89TYY216 pKa = 7.8PTLVEE221 pKa = 4.09NNVAYY226 pKa = 10.49GNGGKK231 pKa = 10.25GIQVTWSDD239 pKa = 4.09YY240 pKa = 8.98VTVSNNTAYY249 pKa = 10.6HH250 pKa = 6.09NNVDD254 pKa = 4.17LQNDD258 pKa = 4.25GTWRR262 pKa = 11.84GEE264 pKa = 3.59ISNAQSSNNTFVNNIAVADD283 pKa = 4.02PSINSNNTAIDD294 pKa = 3.52NNSYY298 pKa = 11.01GGYY301 pKa = 10.41SNDD304 pKa = 3.04NVVWEE309 pKa = 4.96DD310 pKa = 3.18NVLYY314 pKa = 10.54NGKK317 pKa = 7.58TGQMSASTDD326 pKa = 3.34GGNDD330 pKa = 3.22APSAADD336 pKa = 3.49GNLIGVNPGFVNPANGDD353 pKa = 3.53FSISAGSVAVNLGADD368 pKa = 3.62MDD370 pKa = 4.49GSSSVTSPPTTSTPDD385 pKa = 3.47PVVTPDD391 pKa = 4.04PDD393 pKa = 3.8PVVTPDD399 pKa = 3.97PAPTTPAVEE408 pKa = 3.99NAAPVAHH415 pKa = 7.47DD416 pKa = 5.23DD417 pKa = 4.05INFKK421 pKa = 10.09TSPNAPLILQQSQLLSNDD439 pKa = 2.81SDD441 pKa = 4.33ADD443 pKa = 3.48GDD445 pKa = 4.52ALILTNVVGSTQGSAKK461 pKa = 9.16LTSGNNISFTPEE473 pKa = 3.17SDD475 pKa = 3.36ASGTASVTYY484 pKa = 9.4TVSDD488 pKa = 3.66QNGGTDD494 pKa = 3.03TATAYY499 pKa = 9.98IDD501 pKa = 3.47IVAAAPSGPGGG512 pKa = 3.39
Molecular weight: 52.91 kDa
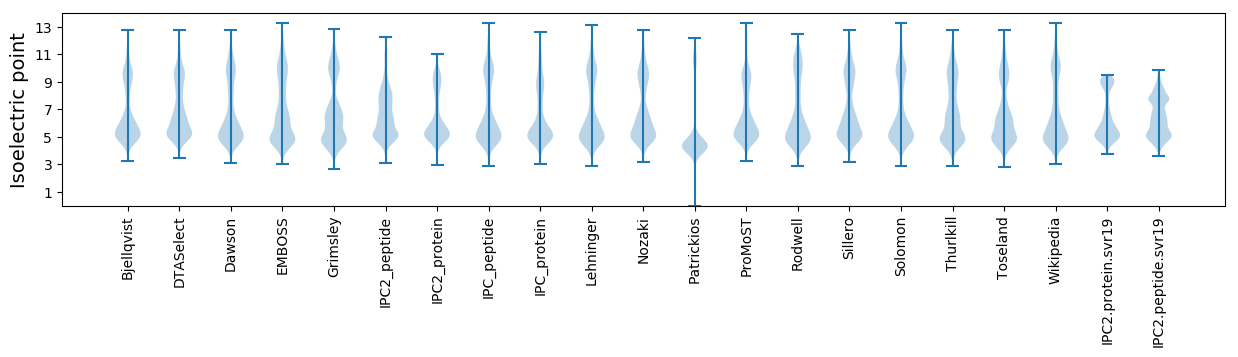
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A074K5E6|A0A074K5E6_9RHOB Uncharacterized protein OS=Thioclava indica OX=1353528 GN=DT23_17610 PE=3 SV=1
MM1 pKa = 7.26AAIVRR6 pKa = 11.84RR7 pKa = 11.84PPAMNTRR14 pKa = 11.84KK15 pKa = 10.33VIGGFPPVFRR25 pKa = 11.84PLTRR29 pKa = 11.84FAFHH33 pKa = 7.33ASRR36 pKa = 11.84FSRR39 pKa = 3.7
MM1 pKa = 7.26AAIVRR6 pKa = 11.84RR7 pKa = 11.84PPAMNTRR14 pKa = 11.84KK15 pKa = 10.33VIGGFPPVFRR25 pKa = 11.84PLTRR29 pKa = 11.84FAFHH33 pKa = 7.33ASRR36 pKa = 11.84FSRR39 pKa = 3.7
Molecular weight: 4.46 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1133096 |
29 |
6803 |
308.7 |
33.45 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.534 ± 0.059 | 0.846 ± 0.015 |
5.916 ± 0.039 | 5.515 ± 0.044 |
3.683 ± 0.033 | 8.624 ± 0.044 |
2.037 ± 0.022 | 5.364 ± 0.032 |
3.523 ± 0.033 | 10.135 ± 0.057 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.84 ± 0.027 | 2.632 ± 0.031 |
5.107 ± 0.033 | 3.386 ± 0.024 |
6.417 ± 0.045 | 5.368 ± 0.047 |
5.376 ± 0.053 | 7.074 ± 0.033 |
1.398 ± 0.019 | 2.225 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
