
Ophiostoma mitovirus 3a
Taxonomy: Viruses; Riboviria; Orthornavirae; Lenarviricota; Howeltoviricetes; Cryppavirales; Mitoviridae; Mitovirus
Average proteome isoelectric point is 8.82
Get precalculated fractions of proteins
Virtual 2D-PAGE plot for 1 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|O92614|O92614_9VIRU RNA-dependent RNA polymerase OS=Ophiostoma mitovirus 3a OX=198597 PE=4 SV=1
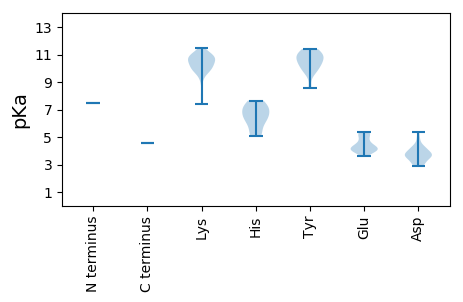
MM1 pKa = 7.48KK2 pKa = 10.55RR3 pKa = 11.84LTLSQNKK10 pKa = 9.7SNQLTNNDD18 pKa = 4.15LSNVGYY24 pKa = 8.54ITKK27 pKa = 10.08QLFPHH32 pKa = 7.65WIRR35 pKa = 11.84LLVWSLQLSPAPYY48 pKa = 9.7KK49 pKa = 10.84KK50 pKa = 10.03FGSRR54 pKa = 11.84IAILWKK60 pKa = 10.51ANGVSFTVQYY70 pKa = 10.92LKK72 pKa = 10.4EE73 pKa = 4.06CTRR76 pKa = 11.84IVQHH80 pKa = 6.14FVSGHH85 pKa = 5.08PVFVTDD91 pKa = 4.26VMPIGLAGGLPTIIPGTLRR110 pKa = 11.84TLLRR114 pKa = 11.84SKK116 pKa = 10.73DD117 pKa = 3.25SSTIRR122 pKa = 11.84GVLSTLAVYY131 pKa = 10.52RR132 pKa = 11.84IMKK135 pKa = 7.39MPCVLKK141 pKa = 10.84LEE143 pKa = 5.12SITDD147 pKa = 3.81PFKK150 pKa = 11.15GISDD154 pKa = 4.18TLPKK158 pKa = 10.33SEE160 pKa = 5.29IINGLASLGFEE171 pKa = 4.04IPKK174 pKa = 10.28GRR176 pKa = 11.84SKK178 pKa = 11.0HH179 pKa = 6.3LLTLSNPIIYY189 pKa = 10.25LLSAGPNHH197 pKa = 7.09SISMMGIWKK206 pKa = 9.75DD207 pKa = 2.94IYY209 pKa = 10.61AWYY212 pKa = 10.22VSPLFPTLLSFIGRR226 pKa = 11.84MNRR229 pKa = 11.84GNVLIDD235 pKa = 3.54LLRR238 pKa = 11.84AEE240 pKa = 4.17VSYY243 pKa = 10.82WEE245 pKa = 4.09ATGVKK250 pKa = 9.77PSVSPLDD257 pKa = 3.73LKK259 pKa = 10.98LGKK262 pKa = 10.04LAIKK266 pKa = 10.33EE267 pKa = 3.98EE268 pKa = 4.33AAGKK272 pKa = 10.46ARR274 pKa = 11.84VFAMADD280 pKa = 3.54SITQSVMAPLNSWVFSKK297 pKa = 11.15LKK299 pKa = 10.03DD300 pKa = 3.63LPMDD304 pKa = 3.56GTFNQQAPLNRR315 pKa = 11.84LVQLYY320 pKa = 10.31QDD322 pKa = 4.19GLLHH326 pKa = 7.16DD327 pKa = 4.3VEE329 pKa = 5.37FYY331 pKa = 11.06SYY333 pKa = 10.88DD334 pKa = 3.48LSSATDD340 pKa = 3.86RR341 pKa = 11.84LPMAFQKK348 pKa = 10.73QIISVLFGSKK358 pKa = 9.11FAKK361 pKa = 10.24DD362 pKa = 3.07WATLLVGRR370 pKa = 11.84DD371 pKa = 3.46WYY373 pKa = 11.38LKK375 pKa = 9.9DD376 pKa = 2.87IPYY379 pKa = 9.72RR380 pKa = 11.84YY381 pKa = 9.82SVGQPMGALSSWAMLALSHH400 pKa = 6.46HH401 pKa = 7.02VIVQIAAMRR410 pKa = 11.84VGKK413 pKa = 10.58LPFTNYY419 pKa = 11.0ALLGDD424 pKa = 5.37DD425 pKa = 4.97IVIADD430 pKa = 3.86KK431 pKa = 11.12AVATSYY437 pKa = 11.22HH438 pKa = 6.3MIMTQILGVEE448 pKa = 4.2INLSKK453 pKa = 11.1SLVSNNSFEE462 pKa = 4.16FAKK465 pKa = 10.7RR466 pKa = 11.84LVTMDD471 pKa = 4.04GEE473 pKa = 4.64VSAVGAKK480 pKa = 10.0NLLVALKK487 pKa = 9.84SRR489 pKa = 11.84WGISSVILDD498 pKa = 4.72LYY500 pKa = 11.4NKK502 pKa = 9.86GLALSEE508 pKa = 3.65QDD510 pKa = 2.93LRR512 pKa = 11.84QRR514 pKa = 11.84FSSIPTVSKK523 pKa = 10.83QFGVDD528 pKa = 3.04KK529 pKa = 10.86LLWLVLGPFGFIPSKK544 pKa = 10.89DD545 pKa = 3.44GLSAFMKK552 pKa = 10.44LNRR555 pKa = 11.84SLSLVDD561 pKa = 3.1MHH563 pKa = 7.44ILLSCVDD570 pKa = 3.73EE571 pKa = 5.3AKK573 pKa = 10.62FDD575 pKa = 4.16LDD577 pKa = 3.63KK578 pKa = 10.87KK579 pKa = 8.12TWEE582 pKa = 4.02ANIQEE587 pKa = 4.63TVHH590 pKa = 5.49TLLRR594 pKa = 11.84FGMLSEE600 pKa = 4.0PAGFEE605 pKa = 4.07VFSDD609 pKa = 4.36FTSSPLYY616 pKa = 10.77SFIRR620 pKa = 11.84GQFGNKK626 pKa = 9.32LSALVQDD633 pKa = 3.8KK634 pKa = 10.0PVRR637 pKa = 11.84RR638 pKa = 11.84LIFDD642 pKa = 3.79GPLLHH647 pKa = 7.11FNFYY651 pKa = 10.21TEE653 pKa = 4.87GWCDD657 pKa = 3.24GLMEE661 pKa = 5.21HH662 pKa = 6.59LTKK665 pKa = 10.65KK666 pKa = 9.73IQSDD670 pKa = 3.76SQEE673 pKa = 4.28TVSPSNPFKK682 pKa = 10.31DD683 pKa = 3.82DD684 pKa = 3.57KK685 pKa = 11.51VILPLRR691 pKa = 11.84GNIKK695 pKa = 10.79GIFFKK700 pKa = 10.93HH701 pKa = 5.16VLALMAEE708 pKa = 4.33RR709 pKa = 11.84DD710 pKa = 3.74PATVMRR716 pKa = 11.84WMM718 pKa = 4.56
MM1 pKa = 7.48KK2 pKa = 10.55RR3 pKa = 11.84LTLSQNKK10 pKa = 9.7SNQLTNNDD18 pKa = 4.15LSNVGYY24 pKa = 8.54ITKK27 pKa = 10.08QLFPHH32 pKa = 7.65WIRR35 pKa = 11.84LLVWSLQLSPAPYY48 pKa = 9.7KK49 pKa = 10.84KK50 pKa = 10.03FGSRR54 pKa = 11.84IAILWKK60 pKa = 10.51ANGVSFTVQYY70 pKa = 10.92LKK72 pKa = 10.4EE73 pKa = 4.06CTRR76 pKa = 11.84IVQHH80 pKa = 6.14FVSGHH85 pKa = 5.08PVFVTDD91 pKa = 4.26VMPIGLAGGLPTIIPGTLRR110 pKa = 11.84TLLRR114 pKa = 11.84SKK116 pKa = 10.73DD117 pKa = 3.25SSTIRR122 pKa = 11.84GVLSTLAVYY131 pKa = 10.52RR132 pKa = 11.84IMKK135 pKa = 7.39MPCVLKK141 pKa = 10.84LEE143 pKa = 5.12SITDD147 pKa = 3.81PFKK150 pKa = 11.15GISDD154 pKa = 4.18TLPKK158 pKa = 10.33SEE160 pKa = 5.29IINGLASLGFEE171 pKa = 4.04IPKK174 pKa = 10.28GRR176 pKa = 11.84SKK178 pKa = 11.0HH179 pKa = 6.3LLTLSNPIIYY189 pKa = 10.25LLSAGPNHH197 pKa = 7.09SISMMGIWKK206 pKa = 9.75DD207 pKa = 2.94IYY209 pKa = 10.61AWYY212 pKa = 10.22VSPLFPTLLSFIGRR226 pKa = 11.84MNRR229 pKa = 11.84GNVLIDD235 pKa = 3.54LLRR238 pKa = 11.84AEE240 pKa = 4.17VSYY243 pKa = 10.82WEE245 pKa = 4.09ATGVKK250 pKa = 9.77PSVSPLDD257 pKa = 3.73LKK259 pKa = 10.98LGKK262 pKa = 10.04LAIKK266 pKa = 10.33EE267 pKa = 3.98EE268 pKa = 4.33AAGKK272 pKa = 10.46ARR274 pKa = 11.84VFAMADD280 pKa = 3.54SITQSVMAPLNSWVFSKK297 pKa = 11.15LKK299 pKa = 10.03DD300 pKa = 3.63LPMDD304 pKa = 3.56GTFNQQAPLNRR315 pKa = 11.84LVQLYY320 pKa = 10.31QDD322 pKa = 4.19GLLHH326 pKa = 7.16DD327 pKa = 4.3VEE329 pKa = 5.37FYY331 pKa = 11.06SYY333 pKa = 10.88DD334 pKa = 3.48LSSATDD340 pKa = 3.86RR341 pKa = 11.84LPMAFQKK348 pKa = 10.73QIISVLFGSKK358 pKa = 9.11FAKK361 pKa = 10.24DD362 pKa = 3.07WATLLVGRR370 pKa = 11.84DD371 pKa = 3.46WYY373 pKa = 11.38LKK375 pKa = 9.9DD376 pKa = 2.87IPYY379 pKa = 9.72RR380 pKa = 11.84YY381 pKa = 9.82SVGQPMGALSSWAMLALSHH400 pKa = 6.46HH401 pKa = 7.02VIVQIAAMRR410 pKa = 11.84VGKK413 pKa = 10.58LPFTNYY419 pKa = 11.0ALLGDD424 pKa = 5.37DD425 pKa = 4.97IVIADD430 pKa = 3.86KK431 pKa = 11.12AVATSYY437 pKa = 11.22HH438 pKa = 6.3MIMTQILGVEE448 pKa = 4.2INLSKK453 pKa = 11.1SLVSNNSFEE462 pKa = 4.16FAKK465 pKa = 10.7RR466 pKa = 11.84LVTMDD471 pKa = 4.04GEE473 pKa = 4.64VSAVGAKK480 pKa = 10.0NLLVALKK487 pKa = 9.84SRR489 pKa = 11.84WGISSVILDD498 pKa = 4.72LYY500 pKa = 11.4NKK502 pKa = 9.86GLALSEE508 pKa = 3.65QDD510 pKa = 2.93LRR512 pKa = 11.84QRR514 pKa = 11.84FSSIPTVSKK523 pKa = 10.83QFGVDD528 pKa = 3.04KK529 pKa = 10.86LLWLVLGPFGFIPSKK544 pKa = 10.89DD545 pKa = 3.44GLSAFMKK552 pKa = 10.44LNRR555 pKa = 11.84SLSLVDD561 pKa = 3.1MHH563 pKa = 7.44ILLSCVDD570 pKa = 3.73EE571 pKa = 5.3AKK573 pKa = 10.62FDD575 pKa = 4.16LDD577 pKa = 3.63KK578 pKa = 10.87KK579 pKa = 8.12TWEE582 pKa = 4.02ANIQEE587 pKa = 4.63TVHH590 pKa = 5.49TLLRR594 pKa = 11.84FGMLSEE600 pKa = 4.0PAGFEE605 pKa = 4.07VFSDD609 pKa = 4.36FTSSPLYY616 pKa = 10.77SFIRR620 pKa = 11.84GQFGNKK626 pKa = 9.32LSALVQDD633 pKa = 3.8KK634 pKa = 10.0PVRR637 pKa = 11.84RR638 pKa = 11.84LIFDD642 pKa = 3.79GPLLHH647 pKa = 7.11FNFYY651 pKa = 10.21TEE653 pKa = 4.87GWCDD657 pKa = 3.24GLMEE661 pKa = 5.21HH662 pKa = 6.59LTKK665 pKa = 10.65KK666 pKa = 9.73IQSDD670 pKa = 3.76SQEE673 pKa = 4.28TVSPSNPFKK682 pKa = 10.31DD683 pKa = 3.82DD684 pKa = 3.57KK685 pKa = 11.51VILPLRR691 pKa = 11.84GNIKK695 pKa = 10.79GIFFKK700 pKa = 10.93HH701 pKa = 5.16VLALMAEE708 pKa = 4.33RR709 pKa = 11.84DD710 pKa = 3.74PATVMRR716 pKa = 11.84WMM718 pKa = 4.56
Molecular weight: 80.28 kDa
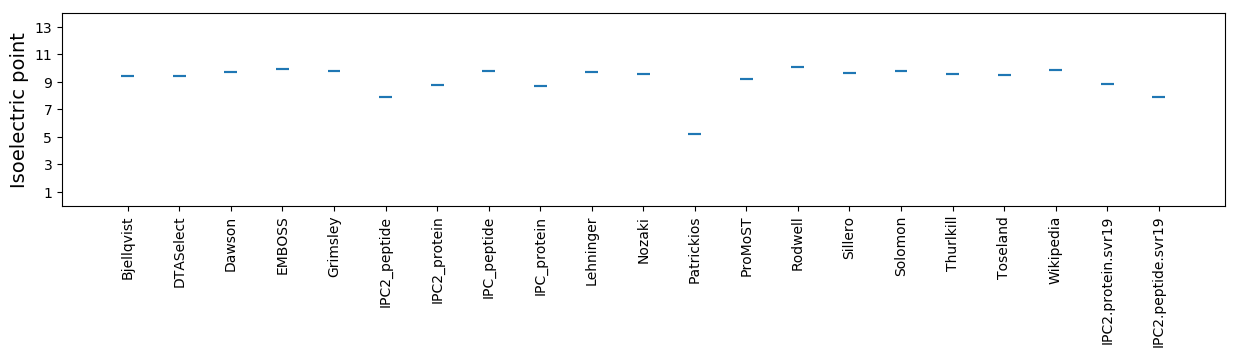
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|O92614|O92614_9VIRU RNA-dependent RNA polymerase OS=Ophiostoma mitovirus 3a OX=198597 PE=4 SV=1
MM1 pKa = 7.48KK2 pKa = 10.55RR3 pKa = 11.84LTLSQNKK10 pKa = 9.7SNQLTNNDD18 pKa = 4.15LSNVGYY24 pKa = 8.54ITKK27 pKa = 10.08QLFPHH32 pKa = 7.65WIRR35 pKa = 11.84LLVWSLQLSPAPYY48 pKa = 9.7KK49 pKa = 10.84KK50 pKa = 10.03FGSRR54 pKa = 11.84IAILWKK60 pKa = 10.51ANGVSFTVQYY70 pKa = 10.92LKK72 pKa = 10.4EE73 pKa = 4.06CTRR76 pKa = 11.84IVQHH80 pKa = 6.14FVSGHH85 pKa = 5.08PVFVTDD91 pKa = 4.26VMPIGLAGGLPTIIPGTLRR110 pKa = 11.84TLLRR114 pKa = 11.84SKK116 pKa = 10.73DD117 pKa = 3.25SSTIRR122 pKa = 11.84GVLSTLAVYY131 pKa = 10.52RR132 pKa = 11.84IMKK135 pKa = 7.39MPCVLKK141 pKa = 10.84LEE143 pKa = 5.12SITDD147 pKa = 3.81PFKK150 pKa = 11.15GISDD154 pKa = 4.18TLPKK158 pKa = 10.33SEE160 pKa = 5.29IINGLASLGFEE171 pKa = 4.04IPKK174 pKa = 10.28GRR176 pKa = 11.84SKK178 pKa = 11.0HH179 pKa = 6.3LLTLSNPIIYY189 pKa = 10.25LLSAGPNHH197 pKa = 7.09SISMMGIWKK206 pKa = 9.75DD207 pKa = 2.94IYY209 pKa = 10.61AWYY212 pKa = 10.22VSPLFPTLLSFIGRR226 pKa = 11.84MNRR229 pKa = 11.84GNVLIDD235 pKa = 3.54LLRR238 pKa = 11.84AEE240 pKa = 4.17VSYY243 pKa = 10.82WEE245 pKa = 4.09ATGVKK250 pKa = 9.77PSVSPLDD257 pKa = 3.73LKK259 pKa = 10.98LGKK262 pKa = 10.04LAIKK266 pKa = 10.33EE267 pKa = 3.98EE268 pKa = 4.33AAGKK272 pKa = 10.46ARR274 pKa = 11.84VFAMADD280 pKa = 3.54SITQSVMAPLNSWVFSKK297 pKa = 11.15LKK299 pKa = 10.03DD300 pKa = 3.63LPMDD304 pKa = 3.56GTFNQQAPLNRR315 pKa = 11.84LVQLYY320 pKa = 10.31QDD322 pKa = 4.19GLLHH326 pKa = 7.16DD327 pKa = 4.3VEE329 pKa = 5.37FYY331 pKa = 11.06SYY333 pKa = 10.88DD334 pKa = 3.48LSSATDD340 pKa = 3.86RR341 pKa = 11.84LPMAFQKK348 pKa = 10.73QIISVLFGSKK358 pKa = 9.11FAKK361 pKa = 10.24DD362 pKa = 3.07WATLLVGRR370 pKa = 11.84DD371 pKa = 3.46WYY373 pKa = 11.38LKK375 pKa = 9.9DD376 pKa = 2.87IPYY379 pKa = 9.72RR380 pKa = 11.84YY381 pKa = 9.82SVGQPMGALSSWAMLALSHH400 pKa = 6.46HH401 pKa = 7.02VIVQIAAMRR410 pKa = 11.84VGKK413 pKa = 10.58LPFTNYY419 pKa = 11.0ALLGDD424 pKa = 5.37DD425 pKa = 4.97IVIADD430 pKa = 3.86KK431 pKa = 11.12AVATSYY437 pKa = 11.22HH438 pKa = 6.3MIMTQILGVEE448 pKa = 4.2INLSKK453 pKa = 11.1SLVSNNSFEE462 pKa = 4.16FAKK465 pKa = 10.7RR466 pKa = 11.84LVTMDD471 pKa = 4.04GEE473 pKa = 4.64VSAVGAKK480 pKa = 10.0NLLVALKK487 pKa = 9.84SRR489 pKa = 11.84WGISSVILDD498 pKa = 4.72LYY500 pKa = 11.4NKK502 pKa = 9.86GLALSEE508 pKa = 3.65QDD510 pKa = 2.93LRR512 pKa = 11.84QRR514 pKa = 11.84FSSIPTVSKK523 pKa = 10.83QFGVDD528 pKa = 3.04KK529 pKa = 10.86LLWLVLGPFGFIPSKK544 pKa = 10.89DD545 pKa = 3.44GLSAFMKK552 pKa = 10.44LNRR555 pKa = 11.84SLSLVDD561 pKa = 3.1MHH563 pKa = 7.44ILLSCVDD570 pKa = 3.73EE571 pKa = 5.3AKK573 pKa = 10.62FDD575 pKa = 4.16LDD577 pKa = 3.63KK578 pKa = 10.87KK579 pKa = 8.12TWEE582 pKa = 4.02ANIQEE587 pKa = 4.63TVHH590 pKa = 5.49TLLRR594 pKa = 11.84FGMLSEE600 pKa = 4.0PAGFEE605 pKa = 4.07VFSDD609 pKa = 4.36FTSSPLYY616 pKa = 10.77SFIRR620 pKa = 11.84GQFGNKK626 pKa = 9.32LSALVQDD633 pKa = 3.8KK634 pKa = 10.0PVRR637 pKa = 11.84RR638 pKa = 11.84LIFDD642 pKa = 3.79GPLLHH647 pKa = 7.11FNFYY651 pKa = 10.21TEE653 pKa = 4.87GWCDD657 pKa = 3.24GLMEE661 pKa = 5.21HH662 pKa = 6.59LTKK665 pKa = 10.65KK666 pKa = 9.73IQSDD670 pKa = 3.76SQEE673 pKa = 4.28TVSPSNPFKK682 pKa = 10.31DD683 pKa = 3.82DD684 pKa = 3.57KK685 pKa = 11.51VILPLRR691 pKa = 11.84GNIKK695 pKa = 10.79GIFFKK700 pKa = 10.93HH701 pKa = 5.16VLALMAEE708 pKa = 4.33RR709 pKa = 11.84DD710 pKa = 3.74PATVMRR716 pKa = 11.84WMM718 pKa = 4.56
MM1 pKa = 7.48KK2 pKa = 10.55RR3 pKa = 11.84LTLSQNKK10 pKa = 9.7SNQLTNNDD18 pKa = 4.15LSNVGYY24 pKa = 8.54ITKK27 pKa = 10.08QLFPHH32 pKa = 7.65WIRR35 pKa = 11.84LLVWSLQLSPAPYY48 pKa = 9.7KK49 pKa = 10.84KK50 pKa = 10.03FGSRR54 pKa = 11.84IAILWKK60 pKa = 10.51ANGVSFTVQYY70 pKa = 10.92LKK72 pKa = 10.4EE73 pKa = 4.06CTRR76 pKa = 11.84IVQHH80 pKa = 6.14FVSGHH85 pKa = 5.08PVFVTDD91 pKa = 4.26VMPIGLAGGLPTIIPGTLRR110 pKa = 11.84TLLRR114 pKa = 11.84SKK116 pKa = 10.73DD117 pKa = 3.25SSTIRR122 pKa = 11.84GVLSTLAVYY131 pKa = 10.52RR132 pKa = 11.84IMKK135 pKa = 7.39MPCVLKK141 pKa = 10.84LEE143 pKa = 5.12SITDD147 pKa = 3.81PFKK150 pKa = 11.15GISDD154 pKa = 4.18TLPKK158 pKa = 10.33SEE160 pKa = 5.29IINGLASLGFEE171 pKa = 4.04IPKK174 pKa = 10.28GRR176 pKa = 11.84SKK178 pKa = 11.0HH179 pKa = 6.3LLTLSNPIIYY189 pKa = 10.25LLSAGPNHH197 pKa = 7.09SISMMGIWKK206 pKa = 9.75DD207 pKa = 2.94IYY209 pKa = 10.61AWYY212 pKa = 10.22VSPLFPTLLSFIGRR226 pKa = 11.84MNRR229 pKa = 11.84GNVLIDD235 pKa = 3.54LLRR238 pKa = 11.84AEE240 pKa = 4.17VSYY243 pKa = 10.82WEE245 pKa = 4.09ATGVKK250 pKa = 9.77PSVSPLDD257 pKa = 3.73LKK259 pKa = 10.98LGKK262 pKa = 10.04LAIKK266 pKa = 10.33EE267 pKa = 3.98EE268 pKa = 4.33AAGKK272 pKa = 10.46ARR274 pKa = 11.84VFAMADD280 pKa = 3.54SITQSVMAPLNSWVFSKK297 pKa = 11.15LKK299 pKa = 10.03DD300 pKa = 3.63LPMDD304 pKa = 3.56GTFNQQAPLNRR315 pKa = 11.84LVQLYY320 pKa = 10.31QDD322 pKa = 4.19GLLHH326 pKa = 7.16DD327 pKa = 4.3VEE329 pKa = 5.37FYY331 pKa = 11.06SYY333 pKa = 10.88DD334 pKa = 3.48LSSATDD340 pKa = 3.86RR341 pKa = 11.84LPMAFQKK348 pKa = 10.73QIISVLFGSKK358 pKa = 9.11FAKK361 pKa = 10.24DD362 pKa = 3.07WATLLVGRR370 pKa = 11.84DD371 pKa = 3.46WYY373 pKa = 11.38LKK375 pKa = 9.9DD376 pKa = 2.87IPYY379 pKa = 9.72RR380 pKa = 11.84YY381 pKa = 9.82SVGQPMGALSSWAMLALSHH400 pKa = 6.46HH401 pKa = 7.02VIVQIAAMRR410 pKa = 11.84VGKK413 pKa = 10.58LPFTNYY419 pKa = 11.0ALLGDD424 pKa = 5.37DD425 pKa = 4.97IVIADD430 pKa = 3.86KK431 pKa = 11.12AVATSYY437 pKa = 11.22HH438 pKa = 6.3MIMTQILGVEE448 pKa = 4.2INLSKK453 pKa = 11.1SLVSNNSFEE462 pKa = 4.16FAKK465 pKa = 10.7RR466 pKa = 11.84LVTMDD471 pKa = 4.04GEE473 pKa = 4.64VSAVGAKK480 pKa = 10.0NLLVALKK487 pKa = 9.84SRR489 pKa = 11.84WGISSVILDD498 pKa = 4.72LYY500 pKa = 11.4NKK502 pKa = 9.86GLALSEE508 pKa = 3.65QDD510 pKa = 2.93LRR512 pKa = 11.84QRR514 pKa = 11.84FSSIPTVSKK523 pKa = 10.83QFGVDD528 pKa = 3.04KK529 pKa = 10.86LLWLVLGPFGFIPSKK544 pKa = 10.89DD545 pKa = 3.44GLSAFMKK552 pKa = 10.44LNRR555 pKa = 11.84SLSLVDD561 pKa = 3.1MHH563 pKa = 7.44ILLSCVDD570 pKa = 3.73EE571 pKa = 5.3AKK573 pKa = 10.62FDD575 pKa = 4.16LDD577 pKa = 3.63KK578 pKa = 10.87KK579 pKa = 8.12TWEE582 pKa = 4.02ANIQEE587 pKa = 4.63TVHH590 pKa = 5.49TLLRR594 pKa = 11.84FGMLSEE600 pKa = 4.0PAGFEE605 pKa = 4.07VFSDD609 pKa = 4.36FTSSPLYY616 pKa = 10.77SFIRR620 pKa = 11.84GQFGNKK626 pKa = 9.32LSALVQDD633 pKa = 3.8KK634 pKa = 10.0PVRR637 pKa = 11.84RR638 pKa = 11.84LIFDD642 pKa = 3.79GPLLHH647 pKa = 7.11FNFYY651 pKa = 10.21TEE653 pKa = 4.87GWCDD657 pKa = 3.24GLMEE661 pKa = 5.21HH662 pKa = 6.59LTKK665 pKa = 10.65KK666 pKa = 9.73IQSDD670 pKa = 3.76SQEE673 pKa = 4.28TVSPSNPFKK682 pKa = 10.31DD683 pKa = 3.82DD684 pKa = 3.57KK685 pKa = 11.51VILPLRR691 pKa = 11.84GNIKK695 pKa = 10.79GIFFKK700 pKa = 10.93HH701 pKa = 5.16VLALMAEE708 pKa = 4.33RR709 pKa = 11.84DD710 pKa = 3.74PATVMRR716 pKa = 11.84WMM718 pKa = 4.56
Molecular weight: 80.28 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
718 |
718 |
718 |
718.0 |
80.28 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.128 ± 0.0 | 0.557 ± 0.0 |
5.292 ± 0.0 | 3.064 ± 0.0 |
5.153 ± 0.0 | 6.407 ± 0.0 |
1.95 ± 0.0 | 6.407 ± 0.0 |
6.685 ± 0.0 | 13.37 ± 0.0 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.343 ± 0.0 | 3.621 ± 0.0 |
4.875 ± 0.0 | 3.343 ± 0.0 |
4.178 ± 0.0 | 9.331 ± 0.0 |
4.596 ± 0.0 | 6.964 ± 0.0 |
2.089 ± 0.0 | 2.646 ± 0.0 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
