
Chryseobacterium sp. 3008163
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Weeksellaceae; Chryseobacterium group; Chryseobacterium; unclassified Chryseobacterium
Average proteome isoelectric point is 6.69
Get precalculated fractions of proteins
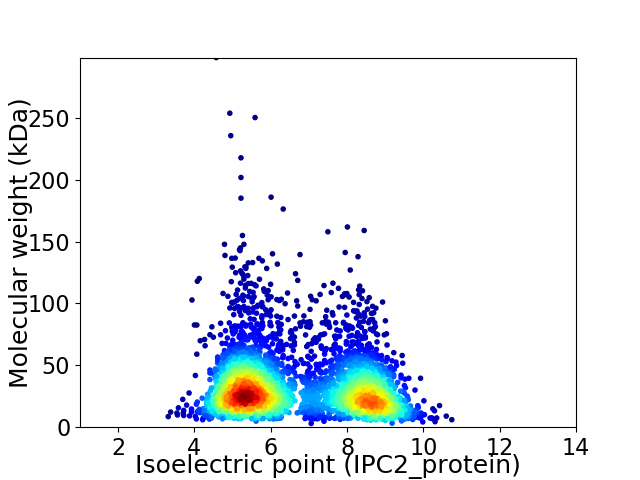
Virtual 2D-PAGE plot for 3789 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
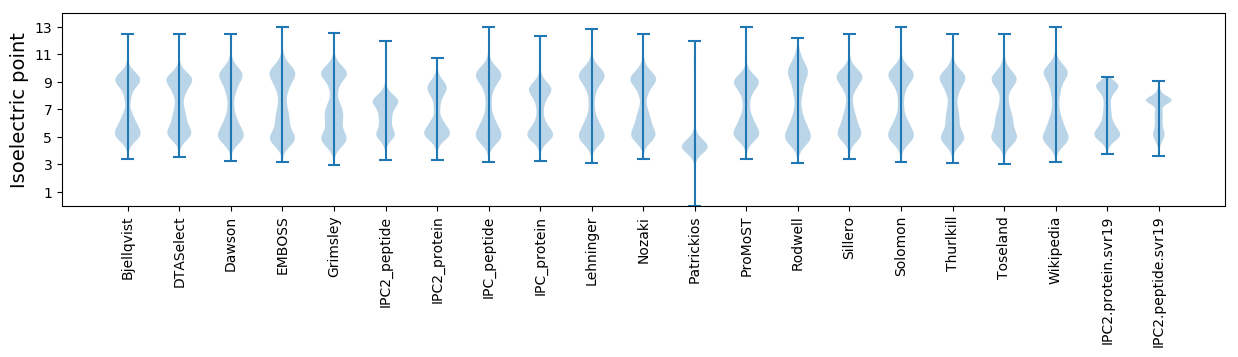
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3G2G9Q3|A0A3G2G9Q3_9FLAO Thioredoxin family protein OS=Chryseobacterium sp. 3008163 OX=2478663 GN=EAG08_10760 PE=4 SV=1
MM1 pKa = 7.39NSYY4 pKa = 9.92IKK6 pKa = 9.86KK7 pKa = 10.11IKK9 pKa = 8.71MKK11 pKa = 10.66KK12 pKa = 10.21NIFKK16 pKa = 10.52YY17 pKa = 10.41VLVPTMILCAFGINFFKK34 pKa = 10.39AQNWTTVWTEE44 pKa = 3.66NFNGLSYY51 pKa = 10.9SSSYY55 pKa = 10.77NNGVTGYY62 pKa = 9.34WNNSANPPQNSQINGAFEE80 pKa = 4.97ANPANNIVAGTTQVPSDD97 pKa = 3.43AAGGRR102 pKa = 11.84FLMFWTQGGVAIPASEE118 pKa = 3.71NVFFKK123 pKa = 10.22KK124 pKa = 9.36TMTVVPGKK132 pKa = 8.39TYY134 pKa = 10.29RR135 pKa = 11.84INYY138 pKa = 8.99RR139 pKa = 11.84FATLGAVPGTATNRR153 pKa = 11.84ANATFRR159 pKa = 11.84VSPQGNTTSFYY170 pKa = 8.76TSGMVVADD178 pKa = 3.32VNAWKK183 pKa = 10.35NVQYY187 pKa = 10.67EE188 pKa = 4.15FTVPSSVTSVDD199 pKa = 3.61MTWEE203 pKa = 4.07NTTKK207 pKa = 9.8STTGNDD213 pKa = 3.23FALDD217 pKa = 3.75DD218 pKa = 4.61LVLQEE223 pKa = 5.55AVDD226 pKa = 3.93SDD228 pKa = 4.03GDD230 pKa = 4.26GIPDD234 pKa = 4.85YY235 pKa = 11.2IDD237 pKa = 6.02DD238 pKa = 5.7DD239 pKa = 4.86DD240 pKa = 6.7DD241 pKa = 5.72NDD243 pKa = 5.25GILDD247 pKa = 3.72TAEE250 pKa = 4.44CGGGAIYY257 pKa = 10.22NYY259 pKa = 10.23SGFGIRR265 pKa = 11.84TISATQANLIGLTGNTYY282 pKa = 9.93TPTTIATNIIPGNDD296 pKa = 3.76ANHH299 pKa = 5.76MTTDD303 pKa = 3.48AARR306 pKa = 11.84NRR308 pKa = 11.84VLFANNIAQGSLFAYY323 pKa = 9.55QFSTNTVVNVGSGFLSSLGDD343 pKa = 3.62ASGGAAMYY351 pKa = 10.92NNDD354 pKa = 3.19YY355 pKa = 10.8FIYY358 pKa = 10.15DD359 pKa = 3.65DD360 pKa = 4.52TGTTSQGLWRR370 pKa = 11.84VTFNASGNASTLTKK384 pKa = 10.54VVDD387 pKa = 4.85PISGIDD393 pKa = 3.86LGDD396 pKa = 3.25IAINSSGMVYY406 pKa = 10.56VIGSTGALYY415 pKa = 10.86KK416 pKa = 10.55LDD418 pKa = 4.24LSSLNLSITTPSSAWISVGTTASASGTQLFFGANGDD454 pKa = 4.44LIGSNSGNLIRR465 pKa = 11.84IDD467 pKa = 3.78PNNAANLGTISTLTSGYY484 pKa = 9.18SWGDD488 pKa = 2.92ISEE491 pKa = 4.36APSVGFSCGTDD502 pKa = 3.13TDD504 pKa = 4.16SDD506 pKa = 4.6GIPNNLDD513 pKa = 3.53LDD515 pKa = 4.21SDD517 pKa = 4.41GDD519 pKa = 3.94GCPDD523 pKa = 3.19ARR525 pKa = 11.84EE526 pKa = 4.1GAGNFNPTTIASGTIASQTPNTNFGTAVSVTTGVPTAVGAGQSVGQSQDD575 pKa = 2.75SSKK578 pKa = 11.24NDD580 pKa = 3.64CLDD583 pKa = 3.32SDD585 pKa = 5.08GDD587 pKa = 5.0GLPDD591 pKa = 3.7WQDD594 pKa = 3.56LDD596 pKa = 4.64DD597 pKa = 6.11DD598 pKa = 4.68NDD600 pKa = 5.0GILDD604 pKa = 3.97TVEE607 pKa = 4.12CTPTYY612 pKa = 10.36LVRR615 pKa = 11.84PVTSSSVTADD625 pKa = 3.18KK626 pKa = 10.73PITVGTAPQIADD638 pKa = 3.55GEE640 pKa = 4.76GAGGSGDD647 pKa = 3.69GPFPYY652 pKa = 9.71WYY654 pKa = 9.14TNVPNLPIAFSMNMQSSSTIDD675 pKa = 3.22HH676 pKa = 6.16VKK678 pKa = 10.65LYY680 pKa = 10.71GPWGFNEE687 pKa = 4.35WIGNFTIEE695 pKa = 5.21LYY697 pKa = 10.8NAGNTLLGTEE707 pKa = 4.43NFVAPDD713 pKa = 3.69QYY715 pKa = 10.88TGTPIFSFTKK725 pKa = 9.8EE726 pKa = 3.9YY727 pKa = 10.73TNVARR732 pKa = 11.84VRR734 pKa = 11.84FTIVSSQGYY743 pKa = 7.73STVTPPRR750 pKa = 11.84ASVNEE755 pKa = 3.9IVFLDD760 pKa = 4.29LQPLTCDD767 pKa = 3.61TDD769 pKa = 3.56NDD771 pKa = 5.2GIPNHH776 pKa = 7.09LDD778 pKa = 3.34LDD780 pKa = 4.19SDD782 pKa = 4.4NDD784 pKa = 3.88GCLDD788 pKa = 4.14AMEE791 pKa = 5.03GDD793 pKa = 4.01EE794 pKa = 4.19NVTVNMLVNAASGLSVGTGSSASNQNLCASGTCVNANGVPTQVNSGGAADD844 pKa = 3.6IGGDD848 pKa = 3.43VGQGIGDD855 pKa = 3.89SQNALISSGCYY866 pKa = 8.82CYY868 pKa = 10.6KK869 pKa = 10.43PEE871 pKa = 4.22VVVAGNPNPVKK882 pKa = 10.7HH883 pKa = 6.72GITALKK889 pKa = 9.5RR890 pKa = 11.84ANSGTSDD897 pKa = 3.32WPVVRR902 pKa = 11.84QSAWTVLEE910 pKa = 4.27SKK912 pKa = 9.68TKK914 pKa = 10.27GFVLNRR920 pKa = 11.84MAFVDD925 pKa = 3.9ADD927 pKa = 4.29SNTATPTTPPLASIPAANYY946 pKa = 8.55VEE948 pKa = 4.61GMMVYY953 pKa = 9.04DD954 pKa = 4.7TVAKK958 pKa = 10.09CLKK961 pKa = 9.76IYY963 pKa = 10.91NGTIWSCFSTQTCPDD978 pKa = 3.14
MM1 pKa = 7.39NSYY4 pKa = 9.92IKK6 pKa = 9.86KK7 pKa = 10.11IKK9 pKa = 8.71MKK11 pKa = 10.66KK12 pKa = 10.21NIFKK16 pKa = 10.52YY17 pKa = 10.41VLVPTMILCAFGINFFKK34 pKa = 10.39AQNWTTVWTEE44 pKa = 3.66NFNGLSYY51 pKa = 10.9SSSYY55 pKa = 10.77NNGVTGYY62 pKa = 9.34WNNSANPPQNSQINGAFEE80 pKa = 4.97ANPANNIVAGTTQVPSDD97 pKa = 3.43AAGGRR102 pKa = 11.84FLMFWTQGGVAIPASEE118 pKa = 3.71NVFFKK123 pKa = 10.22KK124 pKa = 9.36TMTVVPGKK132 pKa = 8.39TYY134 pKa = 10.29RR135 pKa = 11.84INYY138 pKa = 8.99RR139 pKa = 11.84FATLGAVPGTATNRR153 pKa = 11.84ANATFRR159 pKa = 11.84VSPQGNTTSFYY170 pKa = 8.76TSGMVVADD178 pKa = 3.32VNAWKK183 pKa = 10.35NVQYY187 pKa = 10.67EE188 pKa = 4.15FTVPSSVTSVDD199 pKa = 3.61MTWEE203 pKa = 4.07NTTKK207 pKa = 9.8STTGNDD213 pKa = 3.23FALDD217 pKa = 3.75DD218 pKa = 4.61LVLQEE223 pKa = 5.55AVDD226 pKa = 3.93SDD228 pKa = 4.03GDD230 pKa = 4.26GIPDD234 pKa = 4.85YY235 pKa = 11.2IDD237 pKa = 6.02DD238 pKa = 5.7DD239 pKa = 4.86DD240 pKa = 6.7DD241 pKa = 5.72NDD243 pKa = 5.25GILDD247 pKa = 3.72TAEE250 pKa = 4.44CGGGAIYY257 pKa = 10.22NYY259 pKa = 10.23SGFGIRR265 pKa = 11.84TISATQANLIGLTGNTYY282 pKa = 9.93TPTTIATNIIPGNDD296 pKa = 3.76ANHH299 pKa = 5.76MTTDD303 pKa = 3.48AARR306 pKa = 11.84NRR308 pKa = 11.84VLFANNIAQGSLFAYY323 pKa = 9.55QFSTNTVVNVGSGFLSSLGDD343 pKa = 3.62ASGGAAMYY351 pKa = 10.92NNDD354 pKa = 3.19YY355 pKa = 10.8FIYY358 pKa = 10.15DD359 pKa = 3.65DD360 pKa = 4.52TGTTSQGLWRR370 pKa = 11.84VTFNASGNASTLTKK384 pKa = 10.54VVDD387 pKa = 4.85PISGIDD393 pKa = 3.86LGDD396 pKa = 3.25IAINSSGMVYY406 pKa = 10.56VIGSTGALYY415 pKa = 10.86KK416 pKa = 10.55LDD418 pKa = 4.24LSSLNLSITTPSSAWISVGTTASASGTQLFFGANGDD454 pKa = 4.44LIGSNSGNLIRR465 pKa = 11.84IDD467 pKa = 3.78PNNAANLGTISTLTSGYY484 pKa = 9.18SWGDD488 pKa = 2.92ISEE491 pKa = 4.36APSVGFSCGTDD502 pKa = 3.13TDD504 pKa = 4.16SDD506 pKa = 4.6GIPNNLDD513 pKa = 3.53LDD515 pKa = 4.21SDD517 pKa = 4.41GDD519 pKa = 3.94GCPDD523 pKa = 3.19ARR525 pKa = 11.84EE526 pKa = 4.1GAGNFNPTTIASGTIASQTPNTNFGTAVSVTTGVPTAVGAGQSVGQSQDD575 pKa = 2.75SSKK578 pKa = 11.24NDD580 pKa = 3.64CLDD583 pKa = 3.32SDD585 pKa = 5.08GDD587 pKa = 5.0GLPDD591 pKa = 3.7WQDD594 pKa = 3.56LDD596 pKa = 4.64DD597 pKa = 6.11DD598 pKa = 4.68NDD600 pKa = 5.0GILDD604 pKa = 3.97TVEE607 pKa = 4.12CTPTYY612 pKa = 10.36LVRR615 pKa = 11.84PVTSSSVTADD625 pKa = 3.18KK626 pKa = 10.73PITVGTAPQIADD638 pKa = 3.55GEE640 pKa = 4.76GAGGSGDD647 pKa = 3.69GPFPYY652 pKa = 9.71WYY654 pKa = 9.14TNVPNLPIAFSMNMQSSSTIDD675 pKa = 3.22HH676 pKa = 6.16VKK678 pKa = 10.65LYY680 pKa = 10.71GPWGFNEE687 pKa = 4.35WIGNFTIEE695 pKa = 5.21LYY697 pKa = 10.8NAGNTLLGTEE707 pKa = 4.43NFVAPDD713 pKa = 3.69QYY715 pKa = 10.88TGTPIFSFTKK725 pKa = 9.8EE726 pKa = 3.9YY727 pKa = 10.73TNVARR732 pKa = 11.84VRR734 pKa = 11.84FTIVSSQGYY743 pKa = 7.73STVTPPRR750 pKa = 11.84ASVNEE755 pKa = 3.9IVFLDD760 pKa = 4.29LQPLTCDD767 pKa = 3.61TDD769 pKa = 3.56NDD771 pKa = 5.2GIPNHH776 pKa = 7.09LDD778 pKa = 3.34LDD780 pKa = 4.19SDD782 pKa = 4.4NDD784 pKa = 3.88GCLDD788 pKa = 4.14AMEE791 pKa = 5.03GDD793 pKa = 4.01EE794 pKa = 4.19NVTVNMLVNAASGLSVGTGSSASNQNLCASGTCVNANGVPTQVNSGGAADD844 pKa = 3.6IGGDD848 pKa = 3.43VGQGIGDD855 pKa = 3.89SQNALISSGCYY866 pKa = 8.82CYY868 pKa = 10.6KK869 pKa = 10.43PEE871 pKa = 4.22VVVAGNPNPVKK882 pKa = 10.7HH883 pKa = 6.72GITALKK889 pKa = 9.5RR890 pKa = 11.84ANSGTSDD897 pKa = 3.32WPVVRR902 pKa = 11.84QSAWTVLEE910 pKa = 4.27SKK912 pKa = 9.68TKK914 pKa = 10.27GFVLNRR920 pKa = 11.84MAFVDD925 pKa = 3.9ADD927 pKa = 4.29SNTATPTTPPLASIPAANYY946 pKa = 8.55VEE948 pKa = 4.61GMMVYY953 pKa = 9.04DD954 pKa = 4.7TVAKK958 pKa = 10.09CLKK961 pKa = 9.76IYY963 pKa = 10.91NGTIWSCFSTQTCPDD978 pKa = 3.14
Molecular weight: 102.77 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3G2G9K5|A0A3G2G9K5_9FLAO Uncharacterized protein OS=Chryseobacterium sp. 3008163 OX=2478663 GN=EAG08_13650 PE=4 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPSEE10 pKa = 3.58RR11 pKa = 11.84KK12 pKa = 9.37KK13 pKa = 10.2RR14 pKa = 11.84NKK16 pKa = 9.71HH17 pKa = 4.09GFRR20 pKa = 11.84EE21 pKa = 4.33RR22 pKa = 11.84MSTPNGRR29 pKa = 11.84RR30 pKa = 11.84VLAARR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 9.98GRR40 pKa = 11.84KK41 pKa = 8.81SLTVSSARR49 pKa = 11.84AKK51 pKa = 10.4RR52 pKa = 3.45
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPSEE10 pKa = 3.58RR11 pKa = 11.84KK12 pKa = 9.37KK13 pKa = 10.2RR14 pKa = 11.84NKK16 pKa = 9.71HH17 pKa = 4.09GFRR20 pKa = 11.84EE21 pKa = 4.33RR22 pKa = 11.84MSTPNGRR29 pKa = 11.84RR30 pKa = 11.84VLAARR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 9.98GRR40 pKa = 11.84KK41 pKa = 8.81SLTVSSARR49 pKa = 11.84AKK51 pKa = 10.4RR52 pKa = 3.45
Molecular weight: 6.07 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1171329 |
25 |
2833 |
309.1 |
34.99 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.004 ± 0.04 | 0.729 ± 0.012 |
5.403 ± 0.03 | 6.56 ± 0.05 |
5.52 ± 0.034 | 6.233 ± 0.044 |
1.637 ± 0.016 | 8.035 ± 0.043 |
8.404 ± 0.046 | 8.966 ± 0.044 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.315 ± 0.02 | 6.677 ± 0.049 |
3.329 ± 0.022 | 3.53 ± 0.024 |
3.069 ± 0.024 | 6.765 ± 0.032 |
5.653 ± 0.044 | 5.933 ± 0.031 |
1.061 ± 0.016 | 4.178 ± 0.033 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
