
Termite associated circular virus 1
Taxonomy: Viruses; unclassified viruses; unclassified DNA viruses; unclassified ssDNA viruses
Average proteome isoelectric point is 7.86
Get precalculated fractions of proteins
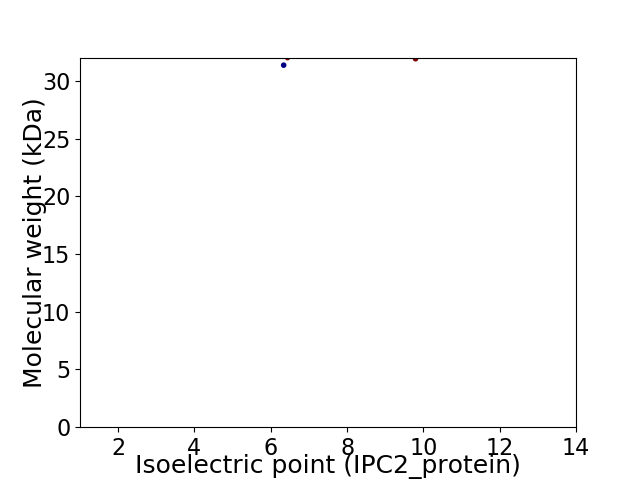
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2P1E327|A0A2P1E327_9VIRU Putative capsid protein OS=Termite associated circular virus 1 OX=2108549 PE=4 SV=1
MM1 pKa = 7.4SFRR4 pKa = 11.84WQGRR8 pKa = 11.84YY9 pKa = 9.71LFLTYY14 pKa = 10.59AQAGNDD20 pKa = 3.57FTPNEE25 pKa = 4.85LITFIRR31 pKa = 11.84EE32 pKa = 4.06SGRR35 pKa = 11.84KK36 pKa = 6.95QPGYY40 pKa = 10.32ILIGKK45 pKa = 7.84EE46 pKa = 3.66RR47 pKa = 11.84HH48 pKa = 5.11QDD50 pKa = 3.17GGLHH54 pKa = 4.6YY55 pKa = 10.43HH56 pKa = 7.07CFVDD60 pKa = 3.52WVKK63 pKa = 10.85RR64 pKa = 11.84FSFTNAARR72 pKa = 11.84FDD74 pKa = 4.26FGGHH78 pKa = 6.44HH79 pKa = 6.9PNISPVNSPKK89 pKa = 10.81ACLDD93 pKa = 3.6YY94 pKa = 10.92CSKK97 pKa = 11.29DD98 pKa = 2.92GDD100 pKa = 4.05TTSWGTPPDD109 pKa = 3.73LDD111 pKa = 4.07SNHH114 pKa = 6.47RR115 pKa = 11.84EE116 pKa = 4.0SRR118 pKa = 11.84NEE120 pKa = 3.38LWSRR124 pKa = 11.84LLDD127 pKa = 3.67TATNPPEE134 pKa = 3.81FLQLVRR140 pKa = 11.84EE141 pKa = 4.11HH142 pKa = 6.84APYY145 pKa = 11.07DD146 pKa = 3.2FATRR150 pKa = 11.84YY151 pKa = 6.46TQLDD155 pKa = 3.43TMARR159 pKa = 11.84AVYY162 pKa = 10.03RR163 pKa = 11.84PRR165 pKa = 11.84TNFTSEE171 pKa = 4.23FEE173 pKa = 4.75HH174 pKa = 7.41EE175 pKa = 5.29DD176 pKa = 3.19FTLPEE181 pKa = 5.12GIEE184 pKa = 4.17GWISSLFTKK193 pKa = 9.87TRR195 pKa = 11.84TAPYY199 pKa = 9.66DD200 pKa = 3.73PSHH203 pKa = 6.9SSSSATLVPEE213 pKa = 4.18RR214 pKa = 11.84PVGRR218 pKa = 11.84GRR220 pKa = 11.84LGTTYY225 pKa = 10.46TSRR228 pKa = 11.84DD229 pKa = 3.31LSTSTNSMPRR239 pKa = 11.84QTTSFSTTAKK249 pKa = 9.62SSSYY253 pKa = 8.22QTTSPGWGAGASSKK267 pKa = 10.2QQTSTEE273 pKa = 4.31EE274 pKa = 4.02KK275 pKa = 9.54EE276 pKa = 4.27HH277 pKa = 6.34
MM1 pKa = 7.4SFRR4 pKa = 11.84WQGRR8 pKa = 11.84YY9 pKa = 9.71LFLTYY14 pKa = 10.59AQAGNDD20 pKa = 3.57FTPNEE25 pKa = 4.85LITFIRR31 pKa = 11.84EE32 pKa = 4.06SGRR35 pKa = 11.84KK36 pKa = 6.95QPGYY40 pKa = 10.32ILIGKK45 pKa = 7.84EE46 pKa = 3.66RR47 pKa = 11.84HH48 pKa = 5.11QDD50 pKa = 3.17GGLHH54 pKa = 4.6YY55 pKa = 10.43HH56 pKa = 7.07CFVDD60 pKa = 3.52WVKK63 pKa = 10.85RR64 pKa = 11.84FSFTNAARR72 pKa = 11.84FDD74 pKa = 4.26FGGHH78 pKa = 6.44HH79 pKa = 6.9PNISPVNSPKK89 pKa = 10.81ACLDD93 pKa = 3.6YY94 pKa = 10.92CSKK97 pKa = 11.29DD98 pKa = 2.92GDD100 pKa = 4.05TTSWGTPPDD109 pKa = 3.73LDD111 pKa = 4.07SNHH114 pKa = 6.47RR115 pKa = 11.84EE116 pKa = 4.0SRR118 pKa = 11.84NEE120 pKa = 3.38LWSRR124 pKa = 11.84LLDD127 pKa = 3.67TATNPPEE134 pKa = 3.81FLQLVRR140 pKa = 11.84EE141 pKa = 4.11HH142 pKa = 6.84APYY145 pKa = 11.07DD146 pKa = 3.2FATRR150 pKa = 11.84YY151 pKa = 6.46TQLDD155 pKa = 3.43TMARR159 pKa = 11.84AVYY162 pKa = 10.03RR163 pKa = 11.84PRR165 pKa = 11.84TNFTSEE171 pKa = 4.23FEE173 pKa = 4.75HH174 pKa = 7.41EE175 pKa = 5.29DD176 pKa = 3.19FTLPEE181 pKa = 5.12GIEE184 pKa = 4.17GWISSLFTKK193 pKa = 9.87TRR195 pKa = 11.84TAPYY199 pKa = 9.66DD200 pKa = 3.73PSHH203 pKa = 6.9SSSSATLVPEE213 pKa = 4.18RR214 pKa = 11.84PVGRR218 pKa = 11.84GRR220 pKa = 11.84LGTTYY225 pKa = 10.46TSRR228 pKa = 11.84DD229 pKa = 3.31LSTSTNSMPRR239 pKa = 11.84QTTSFSTTAKK249 pKa = 9.62SSSYY253 pKa = 8.22QTTSPGWGAGASSKK267 pKa = 10.2QQTSTEE273 pKa = 4.31EE274 pKa = 4.02KK275 pKa = 9.54EE276 pKa = 4.27HH277 pKa = 6.34
Molecular weight: 31.36 kDa
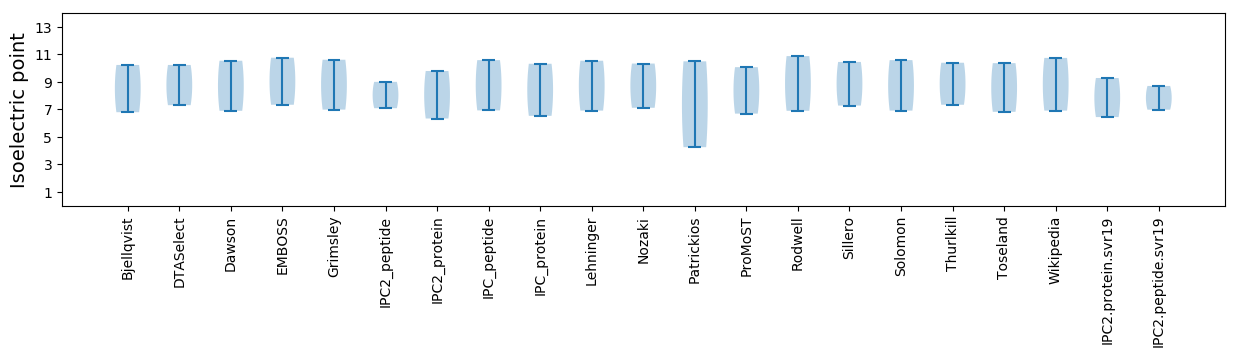
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2P1E327|A0A2P1E327_9VIRU Putative capsid protein OS=Termite associated circular virus 1 OX=2108549 PE=4 SV=1
MM1 pKa = 7.86PYY3 pKa = 10.3SKK5 pKa = 10.16RR6 pKa = 11.84RR7 pKa = 11.84YY8 pKa = 6.51TRR10 pKa = 11.84KK11 pKa = 9.11PYY13 pKa = 10.11RR14 pKa = 11.84RR15 pKa = 11.84STRR18 pKa = 11.84KK19 pKa = 5.8TTTRR23 pKa = 11.84RR24 pKa = 11.84TNRR27 pKa = 11.84RR28 pKa = 11.84RR29 pKa = 11.84SGLTKK34 pKa = 9.26TVARR38 pKa = 11.84ISRR41 pKa = 11.84SVQLRR46 pKa = 11.84QVEE49 pKa = 4.62TKK51 pKa = 10.73RR52 pKa = 11.84NITINLVAAPTPSTNANWQYY72 pKa = 10.2YY73 pKa = 7.34YY74 pKa = 11.23RR75 pKa = 11.84NVFVDD80 pKa = 4.02LANGVDD86 pKa = 3.66SDD88 pKa = 5.33DD89 pKa = 4.27IIGNEE94 pKa = 4.04FVDD97 pKa = 4.6PYY99 pKa = 11.09VTFKK103 pKa = 10.87WNVQFDD109 pKa = 4.14INNLRR114 pKa = 11.84SLNTSNTGVQTVKK127 pKa = 8.15WWMMLVACNDD137 pKa = 3.67YY138 pKa = 11.26LPATVSGFNSIVPGTSTDD156 pKa = 3.41PGGWWVQPDD165 pKa = 3.9PTKK168 pKa = 9.19MTFNGNNVKK177 pKa = 9.84ILKK180 pKa = 10.4KK181 pKa = 7.79MTRR184 pKa = 11.84QWHH187 pKa = 5.27LTSIPSVVGTATVSGTEE204 pKa = 3.93QFSGKK209 pKa = 8.66MKK211 pKa = 9.93YY212 pKa = 8.43RR213 pKa = 11.84WRR215 pKa = 11.84RR216 pKa = 11.84GKK218 pKa = 8.81KK219 pKa = 7.56TFEE222 pKa = 4.1DD223 pKa = 4.0RR224 pKa = 11.84NTIEE228 pKa = 3.73EE229 pKa = 4.23DD230 pKa = 2.99KK231 pKa = 11.16TRR233 pKa = 11.84VSQLRR238 pKa = 11.84GMNYY242 pKa = 10.38YY243 pKa = 9.76IVTGWWVPTALSGTPLAAPIFYY265 pKa = 10.76SVDD268 pKa = 2.82QYY270 pKa = 11.89LYY272 pKa = 10.86FKK274 pKa = 10.98DD275 pKa = 3.63PP276 pKa = 3.56
MM1 pKa = 7.86PYY3 pKa = 10.3SKK5 pKa = 10.16RR6 pKa = 11.84RR7 pKa = 11.84YY8 pKa = 6.51TRR10 pKa = 11.84KK11 pKa = 9.11PYY13 pKa = 10.11RR14 pKa = 11.84RR15 pKa = 11.84STRR18 pKa = 11.84KK19 pKa = 5.8TTTRR23 pKa = 11.84RR24 pKa = 11.84TNRR27 pKa = 11.84RR28 pKa = 11.84RR29 pKa = 11.84SGLTKK34 pKa = 9.26TVARR38 pKa = 11.84ISRR41 pKa = 11.84SVQLRR46 pKa = 11.84QVEE49 pKa = 4.62TKK51 pKa = 10.73RR52 pKa = 11.84NITINLVAAPTPSTNANWQYY72 pKa = 10.2YY73 pKa = 7.34YY74 pKa = 11.23RR75 pKa = 11.84NVFVDD80 pKa = 4.02LANGVDD86 pKa = 3.66SDD88 pKa = 5.33DD89 pKa = 4.27IIGNEE94 pKa = 4.04FVDD97 pKa = 4.6PYY99 pKa = 11.09VTFKK103 pKa = 10.87WNVQFDD109 pKa = 4.14INNLRR114 pKa = 11.84SLNTSNTGVQTVKK127 pKa = 8.15WWMMLVACNDD137 pKa = 3.67YY138 pKa = 11.26LPATVSGFNSIVPGTSTDD156 pKa = 3.41PGGWWVQPDD165 pKa = 3.9PTKK168 pKa = 9.19MTFNGNNVKK177 pKa = 9.84ILKK180 pKa = 10.4KK181 pKa = 7.79MTRR184 pKa = 11.84QWHH187 pKa = 5.27LTSIPSVVGTATVSGTEE204 pKa = 3.93QFSGKK209 pKa = 8.66MKK211 pKa = 9.93YY212 pKa = 8.43RR213 pKa = 11.84WRR215 pKa = 11.84RR216 pKa = 11.84GKK218 pKa = 8.81KK219 pKa = 7.56TFEE222 pKa = 4.1DD223 pKa = 4.0RR224 pKa = 11.84NTIEE228 pKa = 3.73EE229 pKa = 4.23DD230 pKa = 2.99KK231 pKa = 11.16TRR233 pKa = 11.84VSQLRR238 pKa = 11.84GMNYY242 pKa = 10.38YY243 pKa = 9.76IVTGWWVPTALSGTPLAAPIFYY265 pKa = 10.76SVDD268 pKa = 2.82QYY270 pKa = 11.89LYY272 pKa = 10.86FKK274 pKa = 10.98DD275 pKa = 3.63PP276 pKa = 3.56
Molecular weight: 31.92 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
553 |
276 |
277 |
276.5 |
31.64 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.702 ± 0.532 | 0.723 ± 0.268 |
5.063 ± 0.262 | 3.978 ± 1.34 |
4.702 ± 0.801 | 6.329 ± 0.395 |
1.989 ± 1.208 | 3.436 ± 0.677 |
4.702 ± 1.083 | 5.787 ± 0.53 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.808 ± 0.541 | 5.425 ± 1.353 |
5.967 ± 0.396 | 3.617 ± 0.005 |
8.137 ± 0.415 | 9.042 ± 1.602 |
11.573 ± 0.016 | 5.606 ± 2.295 |
2.893 ± 0.542 | 4.521 ± 0.41 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
