
Aedes albopictus densovirus (isolate Boublik/1994) (AalDNV)
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Quintoviricetes; Piccovirales; Parvoviridae; Hamaparvovirinae; Brevihamaparvovirus; Dipteran brevihamaparvovirus 1; Aedes albopictus densovirus
Average proteome isoelectric point is 6.92
Get precalculated fractions of proteins
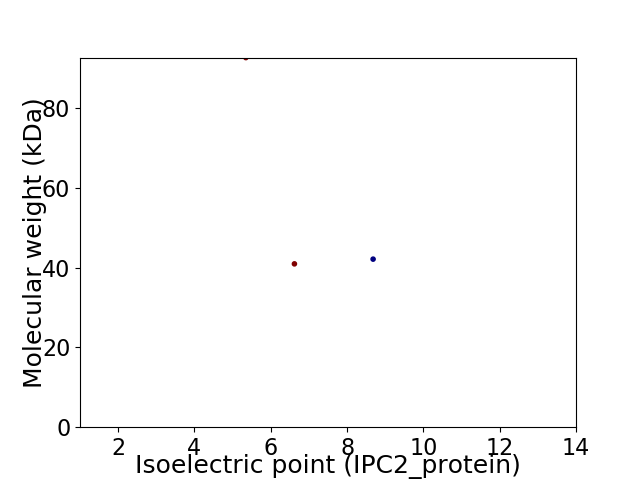
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|Q90186|NS2_AADNV Non-structural protein NS2 OS=Aedes albopictus densovirus (isolate Boublik/1994) OX=648330 GN=NS PE=4 SV=1
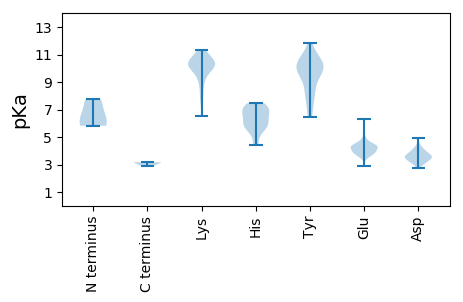
SS1 pKa = 5.82TEE3 pKa = 3.97YY4 pKa = 10.3IRR6 pKa = 11.84GINEE10 pKa = 3.57GRR12 pKa = 11.84VSSMEE17 pKa = 4.32SVCSEE22 pKa = 4.44HH23 pKa = 7.28SPCEE27 pKa = 3.89HH28 pKa = 6.74GNIEE32 pKa = 4.54CEE34 pKa = 5.65CIFCWEE40 pKa = 4.7HH41 pKa = 7.42DD42 pKa = 4.04AQCKK46 pKa = 7.7SRR48 pKa = 11.84RR49 pKa = 11.84RR50 pKa = 11.84EE51 pKa = 3.75LDD53 pKa = 2.87LGEE56 pKa = 4.2PTGSEE61 pKa = 2.89RR62 pKa = 11.84GMANNYY68 pKa = 6.48EE69 pKa = 4.04QSRR72 pKa = 11.84NEE74 pKa = 4.2DD75 pKa = 3.81LYY77 pKa = 10.86CTEE80 pKa = 4.7TVPSSAALQANTITEE95 pKa = 3.79RR96 pKa = 11.84DD97 pKa = 3.27IRR99 pKa = 11.84EE100 pKa = 4.43DD101 pKa = 3.51FTDD104 pKa = 3.38QTVDD108 pKa = 3.18NIYY111 pKa = 9.83PQLHH115 pKa = 5.76SGSRR119 pKa = 11.84ASEE122 pKa = 3.74QLEE125 pKa = 4.25FAFPTIGTRR134 pKa = 11.84SWEE137 pKa = 3.56ILIRR141 pKa = 11.84QSYY144 pKa = 8.5EE145 pKa = 3.71HH146 pKa = 7.02LKK148 pKa = 10.32PDD150 pKa = 3.51YY151 pKa = 10.6KK152 pKa = 11.15EE153 pKa = 4.87EE154 pKa = 4.37DD155 pKa = 3.77FQSHH159 pKa = 4.78IRR161 pKa = 11.84RR162 pKa = 11.84VRR164 pKa = 11.84RR165 pKa = 11.84QLFPEE170 pKa = 3.73KK171 pKa = 10.38TMDD174 pKa = 3.73NNGSQASTTQMLRR187 pKa = 11.84DD188 pKa = 4.48DD189 pKa = 4.38IEE191 pKa = 4.9RR192 pKa = 11.84CGIEE196 pKa = 4.53SIADD200 pKa = 3.53SASEE204 pKa = 4.14DD205 pKa = 3.47NGDD208 pKa = 4.27GVDD211 pKa = 4.02GTCISTVDD219 pKa = 3.26IQGNCIVNAHH229 pKa = 6.2GNEE232 pKa = 3.78QATGSKK238 pKa = 7.63TRR240 pKa = 11.84KK241 pKa = 9.02IRR243 pKa = 11.84ATTTPEE249 pKa = 3.51SSEE252 pKa = 4.31SKK254 pKa = 9.74KK255 pKa = 10.36HH256 pKa = 6.07KK257 pKa = 10.47GSNNQQNIQEE267 pKa = 4.26QSSTTAIADD276 pKa = 3.24GHH278 pKa = 6.84DD279 pKa = 3.61IVDD282 pKa = 3.93GEE284 pKa = 4.32LDD286 pKa = 3.52GRR288 pKa = 11.84SEE290 pKa = 4.57SNRR293 pKa = 11.84EE294 pKa = 3.24TAYY297 pKa = 9.69YY298 pKa = 10.31TFVLHH303 pKa = 7.26KK304 pKa = 10.43NNCKK308 pKa = 10.21EE309 pKa = 3.66DD310 pKa = 2.98WRR312 pKa = 11.84YY313 pKa = 9.85IATTRR318 pKa = 11.84AKK320 pKa = 9.79QAPSFITFDD329 pKa = 3.72HH330 pKa = 7.1GDD332 pKa = 3.85HH333 pKa = 5.75IHH335 pKa = 6.83ILFSSSNTGGNSTRR349 pKa = 11.84VRR351 pKa = 11.84TRR353 pKa = 11.84ITKK356 pKa = 9.82FLSATSAGNAEE367 pKa = 3.98ATITFSKK374 pKa = 11.0VKK376 pKa = 10.19FLRR379 pKa = 11.84NYY381 pKa = 9.03ILYY384 pKa = 9.52CIRR387 pKa = 11.84YY388 pKa = 9.33GIEE391 pKa = 3.76TVNIYY396 pKa = 10.54GNKK399 pKa = 9.09IQQQLTEE406 pKa = 4.25AMDD409 pKa = 3.61TFKK412 pKa = 11.29VLFEE416 pKa = 4.42NRR418 pKa = 11.84DD419 pKa = 3.73PNDD422 pKa = 3.43VILDD426 pKa = 4.19AGCKK430 pKa = 9.61LYY432 pKa = 10.86HH433 pKa = 6.09EE434 pKa = 4.93EE435 pKa = 4.4KK436 pKa = 10.6KK437 pKa = 10.6EE438 pKa = 3.89YY439 pKa = 7.85QQKK442 pKa = 10.17RR443 pKa = 11.84CGQRR447 pKa = 11.84KK448 pKa = 6.52QQNLTDD454 pKa = 4.96IILDD458 pKa = 3.79KK459 pKa = 10.92IKK461 pKa = 10.44EE462 pKa = 4.23KK463 pKa = 10.78KK464 pKa = 8.38ITTAQQWEE472 pKa = 4.47NIIEE476 pKa = 4.16PEE478 pKa = 4.67FKK480 pKa = 10.12IQLMKK485 pKa = 10.94EE486 pKa = 3.78FGLNVDD492 pKa = 4.19SYY494 pKa = 8.03VQRR497 pKa = 11.84IVRR500 pKa = 11.84IEE502 pKa = 3.78RR503 pKa = 11.84TRR505 pKa = 11.84IQQIIKK511 pKa = 10.43SKK513 pKa = 8.69TLTEE517 pKa = 3.93IMVEE521 pKa = 3.97ILNEE525 pKa = 3.96EE526 pKa = 4.45YY527 pKa = 10.31IKK529 pKa = 11.16NFTPGEE535 pKa = 4.33DD536 pKa = 3.23NSKK539 pKa = 10.05LQNIIQWIEE548 pKa = 3.64YY549 pKa = 8.05MFKK552 pKa = 10.64EE553 pKa = 3.86NNINIIHH560 pKa = 6.27FLAWNEE566 pKa = 3.68IIKK569 pKa = 9.87PKK571 pKa = 10.06RR572 pKa = 11.84YY573 pKa = 9.83KK574 pKa = 10.38KK575 pKa = 9.96INGMVLEE582 pKa = 5.46GITNAGKK589 pKa = 10.41SLILDD594 pKa = 3.63NLLAMVKK601 pKa = 10.05PEE603 pKa = 4.45EE604 pKa = 4.29IPRR607 pKa = 11.84EE608 pKa = 3.78RR609 pKa = 11.84DD610 pKa = 2.95NSGFHH615 pKa = 7.45LDD617 pKa = 3.66QLPGAGSVLFEE628 pKa = 4.58EE629 pKa = 5.22PMITPVNVGTWKK641 pKa = 10.82LLLEE645 pKa = 4.64GKK647 pKa = 8.26TIKK650 pKa = 10.01TDD652 pKa = 3.32VKK654 pKa = 11.12NKK656 pKa = 10.07DD657 pKa = 3.23KK658 pKa = 11.17EE659 pKa = 4.56PIEE662 pKa = 4.25RR663 pKa = 11.84TPTWITTATPITNNVHH679 pKa = 5.68MNEE682 pKa = 4.07TSQILQRR689 pKa = 11.84IKK691 pKa = 10.83LYY693 pKa = 10.29IFKK696 pKa = 10.62KK697 pKa = 9.96SIQHH701 pKa = 6.66RR702 pKa = 11.84EE703 pKa = 3.65DD704 pKa = 3.54KK705 pKa = 10.28YY706 pKa = 9.68TINAQIQNKK715 pKa = 9.68LISRR719 pKa = 11.84PPGLVEE725 pKa = 5.08PIHH728 pKa = 5.41MAIVFVKK735 pKa = 10.26NFKK738 pKa = 10.27EE739 pKa = 3.88IYY741 pKa = 9.99KK742 pKa = 10.57LIEE745 pKa = 4.35EE746 pKa = 4.69EE747 pKa = 4.95DD748 pKa = 3.58NAHH751 pKa = 5.59TVNEE755 pKa = 4.01KK756 pKa = 10.55AIRR759 pKa = 11.84LSEE762 pKa = 4.04EE763 pKa = 3.87AKK765 pKa = 10.52QEE767 pKa = 3.98AEE769 pKa = 3.92EE770 pKa = 4.24WQTALQWNTMEE781 pKa = 4.73EE782 pKa = 4.1EE783 pKa = 4.06QKK785 pKa = 11.02EE786 pKa = 4.24NEE788 pKa = 4.49KK789 pKa = 8.88QTEE792 pKa = 4.19DD793 pKa = 3.2QDD795 pKa = 3.99KK796 pKa = 10.63EE797 pKa = 4.53SEE799 pKa = 4.53KK800 pKa = 10.25EE801 pKa = 3.94TATQQ805 pKa = 3.17
SS1 pKa = 5.82TEE3 pKa = 3.97YY4 pKa = 10.3IRR6 pKa = 11.84GINEE10 pKa = 3.57GRR12 pKa = 11.84VSSMEE17 pKa = 4.32SVCSEE22 pKa = 4.44HH23 pKa = 7.28SPCEE27 pKa = 3.89HH28 pKa = 6.74GNIEE32 pKa = 4.54CEE34 pKa = 5.65CIFCWEE40 pKa = 4.7HH41 pKa = 7.42DD42 pKa = 4.04AQCKK46 pKa = 7.7SRR48 pKa = 11.84RR49 pKa = 11.84RR50 pKa = 11.84EE51 pKa = 3.75LDD53 pKa = 2.87LGEE56 pKa = 4.2PTGSEE61 pKa = 2.89RR62 pKa = 11.84GMANNYY68 pKa = 6.48EE69 pKa = 4.04QSRR72 pKa = 11.84NEE74 pKa = 4.2DD75 pKa = 3.81LYY77 pKa = 10.86CTEE80 pKa = 4.7TVPSSAALQANTITEE95 pKa = 3.79RR96 pKa = 11.84DD97 pKa = 3.27IRR99 pKa = 11.84EE100 pKa = 4.43DD101 pKa = 3.51FTDD104 pKa = 3.38QTVDD108 pKa = 3.18NIYY111 pKa = 9.83PQLHH115 pKa = 5.76SGSRR119 pKa = 11.84ASEE122 pKa = 3.74QLEE125 pKa = 4.25FAFPTIGTRR134 pKa = 11.84SWEE137 pKa = 3.56ILIRR141 pKa = 11.84QSYY144 pKa = 8.5EE145 pKa = 3.71HH146 pKa = 7.02LKK148 pKa = 10.32PDD150 pKa = 3.51YY151 pKa = 10.6KK152 pKa = 11.15EE153 pKa = 4.87EE154 pKa = 4.37DD155 pKa = 3.77FQSHH159 pKa = 4.78IRR161 pKa = 11.84RR162 pKa = 11.84VRR164 pKa = 11.84RR165 pKa = 11.84QLFPEE170 pKa = 3.73KK171 pKa = 10.38TMDD174 pKa = 3.73NNGSQASTTQMLRR187 pKa = 11.84DD188 pKa = 4.48DD189 pKa = 4.38IEE191 pKa = 4.9RR192 pKa = 11.84CGIEE196 pKa = 4.53SIADD200 pKa = 3.53SASEE204 pKa = 4.14DD205 pKa = 3.47NGDD208 pKa = 4.27GVDD211 pKa = 4.02GTCISTVDD219 pKa = 3.26IQGNCIVNAHH229 pKa = 6.2GNEE232 pKa = 3.78QATGSKK238 pKa = 7.63TRR240 pKa = 11.84KK241 pKa = 9.02IRR243 pKa = 11.84ATTTPEE249 pKa = 3.51SSEE252 pKa = 4.31SKK254 pKa = 9.74KK255 pKa = 10.36HH256 pKa = 6.07KK257 pKa = 10.47GSNNQQNIQEE267 pKa = 4.26QSSTTAIADD276 pKa = 3.24GHH278 pKa = 6.84DD279 pKa = 3.61IVDD282 pKa = 3.93GEE284 pKa = 4.32LDD286 pKa = 3.52GRR288 pKa = 11.84SEE290 pKa = 4.57SNRR293 pKa = 11.84EE294 pKa = 3.24TAYY297 pKa = 9.69YY298 pKa = 10.31TFVLHH303 pKa = 7.26KK304 pKa = 10.43NNCKK308 pKa = 10.21EE309 pKa = 3.66DD310 pKa = 2.98WRR312 pKa = 11.84YY313 pKa = 9.85IATTRR318 pKa = 11.84AKK320 pKa = 9.79QAPSFITFDD329 pKa = 3.72HH330 pKa = 7.1GDD332 pKa = 3.85HH333 pKa = 5.75IHH335 pKa = 6.83ILFSSSNTGGNSTRR349 pKa = 11.84VRR351 pKa = 11.84TRR353 pKa = 11.84ITKK356 pKa = 9.82FLSATSAGNAEE367 pKa = 3.98ATITFSKK374 pKa = 11.0VKK376 pKa = 10.19FLRR379 pKa = 11.84NYY381 pKa = 9.03ILYY384 pKa = 9.52CIRR387 pKa = 11.84YY388 pKa = 9.33GIEE391 pKa = 3.76TVNIYY396 pKa = 10.54GNKK399 pKa = 9.09IQQQLTEE406 pKa = 4.25AMDD409 pKa = 3.61TFKK412 pKa = 11.29VLFEE416 pKa = 4.42NRR418 pKa = 11.84DD419 pKa = 3.73PNDD422 pKa = 3.43VILDD426 pKa = 4.19AGCKK430 pKa = 9.61LYY432 pKa = 10.86HH433 pKa = 6.09EE434 pKa = 4.93EE435 pKa = 4.4KK436 pKa = 10.6KK437 pKa = 10.6EE438 pKa = 3.89YY439 pKa = 7.85QQKK442 pKa = 10.17RR443 pKa = 11.84CGQRR447 pKa = 11.84KK448 pKa = 6.52QQNLTDD454 pKa = 4.96IILDD458 pKa = 3.79KK459 pKa = 10.92IKK461 pKa = 10.44EE462 pKa = 4.23KK463 pKa = 10.78KK464 pKa = 8.38ITTAQQWEE472 pKa = 4.47NIIEE476 pKa = 4.16PEE478 pKa = 4.67FKK480 pKa = 10.12IQLMKK485 pKa = 10.94EE486 pKa = 3.78FGLNVDD492 pKa = 4.19SYY494 pKa = 8.03VQRR497 pKa = 11.84IVRR500 pKa = 11.84IEE502 pKa = 3.78RR503 pKa = 11.84TRR505 pKa = 11.84IQQIIKK511 pKa = 10.43SKK513 pKa = 8.69TLTEE517 pKa = 3.93IMVEE521 pKa = 3.97ILNEE525 pKa = 3.96EE526 pKa = 4.45YY527 pKa = 10.31IKK529 pKa = 11.16NFTPGEE535 pKa = 4.33DD536 pKa = 3.23NSKK539 pKa = 10.05LQNIIQWIEE548 pKa = 3.64YY549 pKa = 8.05MFKK552 pKa = 10.64EE553 pKa = 3.86NNINIIHH560 pKa = 6.27FLAWNEE566 pKa = 3.68IIKK569 pKa = 9.87PKK571 pKa = 10.06RR572 pKa = 11.84YY573 pKa = 9.83KK574 pKa = 10.38KK575 pKa = 9.96INGMVLEE582 pKa = 5.46GITNAGKK589 pKa = 10.41SLILDD594 pKa = 3.63NLLAMVKK601 pKa = 10.05PEE603 pKa = 4.45EE604 pKa = 4.29IPRR607 pKa = 11.84EE608 pKa = 3.78RR609 pKa = 11.84DD610 pKa = 2.95NSGFHH615 pKa = 7.45LDD617 pKa = 3.66QLPGAGSVLFEE628 pKa = 4.58EE629 pKa = 5.22PMITPVNVGTWKK641 pKa = 10.82LLLEE645 pKa = 4.64GKK647 pKa = 8.26TIKK650 pKa = 10.01TDD652 pKa = 3.32VKK654 pKa = 11.12NKK656 pKa = 10.07DD657 pKa = 3.23KK658 pKa = 11.17EE659 pKa = 4.56PIEE662 pKa = 4.25RR663 pKa = 11.84TPTWITTATPITNNVHH679 pKa = 5.68MNEE682 pKa = 4.07TSQILQRR689 pKa = 11.84IKK691 pKa = 10.83LYY693 pKa = 10.29IFKK696 pKa = 10.62KK697 pKa = 9.96SIQHH701 pKa = 6.66RR702 pKa = 11.84EE703 pKa = 3.65DD704 pKa = 3.54KK705 pKa = 10.28YY706 pKa = 9.68TINAQIQNKK715 pKa = 9.68LISRR719 pKa = 11.84PPGLVEE725 pKa = 5.08PIHH728 pKa = 5.41MAIVFVKK735 pKa = 10.26NFKK738 pKa = 10.27EE739 pKa = 3.88IYY741 pKa = 9.99KK742 pKa = 10.57LIEE745 pKa = 4.35EE746 pKa = 4.69EE747 pKa = 4.95DD748 pKa = 3.58NAHH751 pKa = 5.59TVNEE755 pKa = 4.01KK756 pKa = 10.55AIRR759 pKa = 11.84LSEE762 pKa = 4.04EE763 pKa = 3.87AKK765 pKa = 10.52QEE767 pKa = 3.98AEE769 pKa = 3.92EE770 pKa = 4.24WQTALQWNTMEE781 pKa = 4.73EE782 pKa = 4.1EE783 pKa = 4.06QKK785 pKa = 11.02EE786 pKa = 4.24NEE788 pKa = 4.49KK789 pKa = 8.88QTEE792 pKa = 4.19DD793 pKa = 3.2QDD795 pKa = 3.99KK796 pKa = 10.63EE797 pKa = 4.53SEE799 pKa = 4.53KK800 pKa = 10.25EE801 pKa = 3.94TATQQ805 pKa = 3.17
Molecular weight: 92.64 kDa
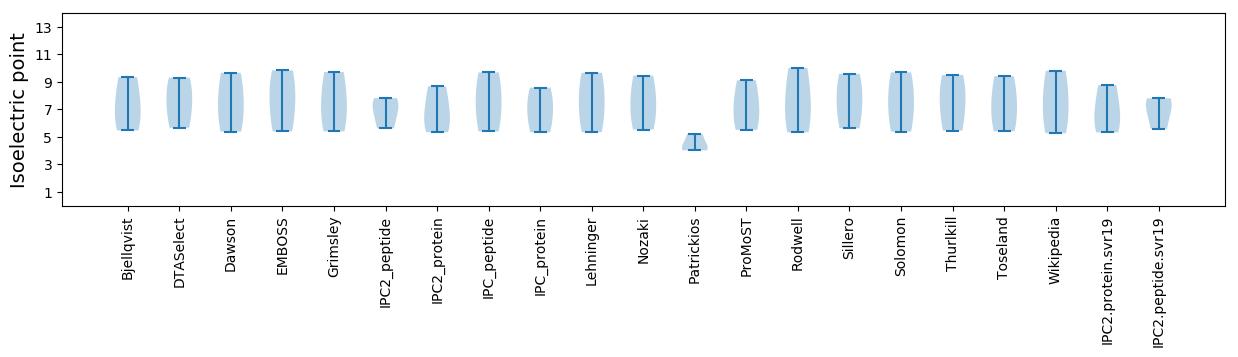
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|Q90187|CAPSD_AADNV Capsid protein VP1/VP2 OS=Aedes albopictus densovirus (isolate Boublik/1994) OX=648330 GN=VP PE=3 SV=2
SS1 pKa = 6.17VSAFFVGSMTHH12 pKa = 5.5SVNPGGEE19 pKa = 3.63NWIWEE24 pKa = 4.36SQLEE28 pKa = 4.31VRR30 pKa = 11.84EE31 pKa = 3.97EE32 pKa = 3.99WPTITNNQGTKK43 pKa = 9.63IYY45 pKa = 9.99IAPRR49 pKa = 11.84QYY51 pKa = 11.42LLQLRR56 pKa = 11.84CKK58 pKa = 7.69QTPSRR63 pKa = 11.84SEE65 pKa = 3.46ISEE68 pKa = 4.45KK69 pKa = 10.28ISQIKK74 pKa = 9.24PLITSTHH81 pKa = 6.52NYY83 pKa = 6.57TAAPEE88 pKa = 4.11PLNSLNSHH96 pKa = 6.1FQLLEE101 pKa = 3.96QEE103 pKa = 4.17AGKK106 pKa = 10.39YY107 pKa = 10.5LYY109 pKa = 11.33VNLTNISSQIIRR121 pKa = 11.84KK122 pKa = 9.75KK123 pKa = 10.2IFNHH127 pKa = 5.81ILDD130 pKa = 4.04VFEE133 pKa = 6.28DD134 pKa = 3.95SYY136 pKa = 11.86SPKK139 pKa = 10.25KK140 pKa = 10.11LWIITGHH147 pKa = 6.09RR148 pKa = 11.84QAQPKK153 pKa = 10.18CYY155 pKa = 7.62EE156 pKa = 4.3TTSKK160 pKa = 11.15DD161 pKa = 3.62AVLKK165 pKa = 10.97ALLTAQAKK173 pKa = 6.97ITEE176 pKa = 4.21MEE178 pKa = 4.15LMGRR182 pKa = 11.84ASPPWIYY189 pKa = 10.74KK190 pKa = 8.82ATVLLTHH197 pKa = 6.7MAMNRR202 pKa = 11.84QQAVRR207 pKa = 11.84PEE209 pKa = 4.31RR210 pKa = 11.84YY211 pKa = 8.67EE212 pKa = 3.62QQPRR216 pKa = 11.84QNQVNQKK223 pKa = 9.35NIKK226 pKa = 9.51AAIINKK232 pKa = 9.35IYY234 pKa = 10.28KK235 pKa = 9.57NKK237 pKa = 10.09AVPPPSPTDD246 pKa = 3.53TISSTEE252 pKa = 3.49SWMEE256 pKa = 3.58EE257 pKa = 3.71VNQIEE262 pKa = 4.56KK263 pKa = 10.01QHH265 pKa = 5.32TTHH268 pKa = 6.99SSSTKK273 pKa = 8.58ITAKK277 pKa = 9.22RR278 pKa = 11.84TGDD281 pKa = 3.15ISPQQGPSKK290 pKa = 10.34RR291 pKa = 11.84RR292 pKa = 11.84ALSPLITEE300 pKa = 4.48TTSTSSSHH308 pKa = 6.37HH309 pKa = 4.39QTQEE313 pKa = 3.36EE314 pKa = 4.54TAQEE318 pKa = 4.31SEE320 pKa = 4.37PEE322 pKa = 4.25SPSFLVQQAQEE333 pKa = 4.0MQKK336 pKa = 10.42RR337 pKa = 11.84LSLFPKK343 pKa = 10.22LNFSGTTFSIASVTVSKK360 pKa = 10.83QSISMEE366 pKa = 3.53IKK368 pKa = 10.69SNNNN372 pKa = 2.93
SS1 pKa = 6.17VSAFFVGSMTHH12 pKa = 5.5SVNPGGEE19 pKa = 3.63NWIWEE24 pKa = 4.36SQLEE28 pKa = 4.31VRR30 pKa = 11.84EE31 pKa = 3.97EE32 pKa = 3.99WPTITNNQGTKK43 pKa = 9.63IYY45 pKa = 9.99IAPRR49 pKa = 11.84QYY51 pKa = 11.42LLQLRR56 pKa = 11.84CKK58 pKa = 7.69QTPSRR63 pKa = 11.84SEE65 pKa = 3.46ISEE68 pKa = 4.45KK69 pKa = 10.28ISQIKK74 pKa = 9.24PLITSTHH81 pKa = 6.52NYY83 pKa = 6.57TAAPEE88 pKa = 4.11PLNSLNSHH96 pKa = 6.1FQLLEE101 pKa = 3.96QEE103 pKa = 4.17AGKK106 pKa = 10.39YY107 pKa = 10.5LYY109 pKa = 11.33VNLTNISSQIIRR121 pKa = 11.84KK122 pKa = 9.75KK123 pKa = 10.2IFNHH127 pKa = 5.81ILDD130 pKa = 4.04VFEE133 pKa = 6.28DD134 pKa = 3.95SYY136 pKa = 11.86SPKK139 pKa = 10.25KK140 pKa = 10.11LWIITGHH147 pKa = 6.09RR148 pKa = 11.84QAQPKK153 pKa = 10.18CYY155 pKa = 7.62EE156 pKa = 4.3TTSKK160 pKa = 11.15DD161 pKa = 3.62AVLKK165 pKa = 10.97ALLTAQAKK173 pKa = 6.97ITEE176 pKa = 4.21MEE178 pKa = 4.15LMGRR182 pKa = 11.84ASPPWIYY189 pKa = 10.74KK190 pKa = 8.82ATVLLTHH197 pKa = 6.7MAMNRR202 pKa = 11.84QQAVRR207 pKa = 11.84PEE209 pKa = 4.31RR210 pKa = 11.84YY211 pKa = 8.67EE212 pKa = 3.62QQPRR216 pKa = 11.84QNQVNQKK223 pKa = 9.35NIKK226 pKa = 9.51AAIINKK232 pKa = 9.35IYY234 pKa = 10.28KK235 pKa = 9.57NKK237 pKa = 10.09AVPPPSPTDD246 pKa = 3.53TISSTEE252 pKa = 3.49SWMEE256 pKa = 3.58EE257 pKa = 3.71VNQIEE262 pKa = 4.56KK263 pKa = 10.01QHH265 pKa = 5.32TTHH268 pKa = 6.99SSSTKK273 pKa = 8.58ITAKK277 pKa = 9.22RR278 pKa = 11.84TGDD281 pKa = 3.15ISPQQGPSKK290 pKa = 10.34RR291 pKa = 11.84RR292 pKa = 11.84ALSPLITEE300 pKa = 4.48TTSTSSSHH308 pKa = 6.37HH309 pKa = 4.39QTQEE313 pKa = 3.36EE314 pKa = 4.54TAQEE318 pKa = 4.31SEE320 pKa = 4.37PEE322 pKa = 4.25SPSFLVQQAQEE333 pKa = 4.0MQKK336 pKa = 10.42RR337 pKa = 11.84LSLFPKK343 pKa = 10.22LNFSGTTFSIASVTVSKK360 pKa = 10.83QSISMEE366 pKa = 3.53IKK368 pKa = 10.69SNNNN372 pKa = 2.93
Molecular weight: 42.13 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1532 |
355 |
805 |
510.7 |
58.57 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.157 ± 0.186 | 1.11 ± 0.436 |
3.851 ± 0.698 | 8.812 ± 1.403 |
3.198 ± 0.391 | 4.634 ± 0.636 |
2.48 ± 0.087 | 8.094 ± 1.312 |
6.527 ± 1.022 | 5.94 ± 0.189 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.35 ± 0.652 | 6.984 ± 0.772 |
4.047 ± 0.599 | 6.658 ± 0.461 |
5.157 ± 0.277 | 7.115 ± 1.273 |
9.138 ± 1.346 | 3.982 ± 0.228 |
1.371 ± 0.072 | 3.394 ± 0.881 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
