
Candida viswanathii
Taxonomy: cellular organisms;
Average proteome isoelectric point is 6.22
Get precalculated fractions of proteins
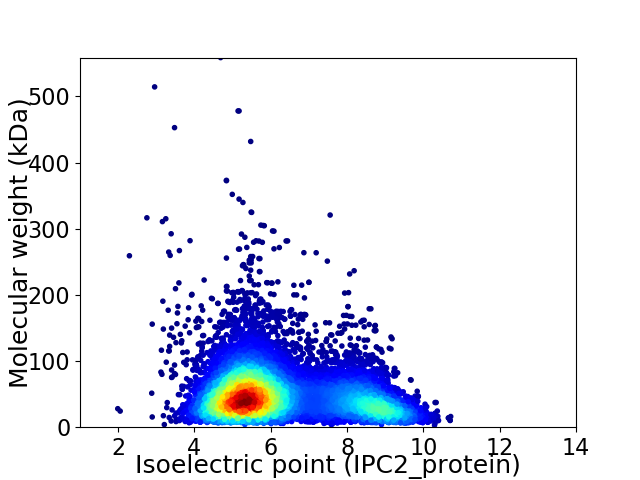
Virtual 2D-PAGE plot for 10576 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A367YFG1|A0A367YFG1_9ASCO Protein BIM1 OS=Candida viswanathii OX=5486 GN=BIM1_1 PE=3 SV=1
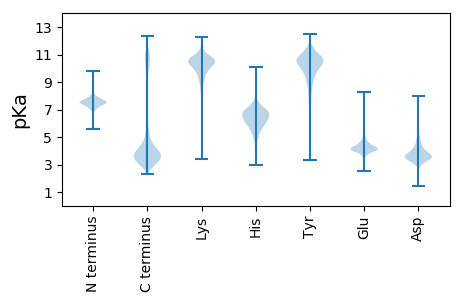
MM1 pKa = 8.11DD2 pKa = 5.97ANLEE6 pKa = 4.05NTKK9 pKa = 8.51MWEE12 pKa = 4.35SVDD15 pKa = 4.21HH16 pKa = 6.71SLTSNLLSRR25 pKa = 11.84ITSTVEE31 pKa = 3.46CTICSEE37 pKa = 3.86IMAVPVTAEE46 pKa = 4.35CGHH49 pKa = 5.91SFCYY53 pKa = 10.36DD54 pKa = 4.35CIYY57 pKa = 10.84QWFSNKK63 pKa = 8.75VNCPTCRR70 pKa = 11.84HH71 pKa = 5.95EE72 pKa = 5.0IEE74 pKa = 4.73NKK76 pKa = 7.93PTLNLQLKK84 pKa = 7.16EE85 pKa = 3.8ICKK88 pKa = 10.25NLLDD92 pKa = 4.43MIIDD96 pKa = 3.67SRR98 pKa = 11.84LEE100 pKa = 3.64NHH102 pKa = 6.46EE103 pKa = 3.84VLKK106 pKa = 10.49NHH108 pKa = 6.66KK109 pKa = 10.24LEE111 pKa = 4.6SEE113 pKa = 4.03NNYY116 pKa = 10.32NLDD119 pKa = 3.33VRR121 pKa = 11.84NKK123 pKa = 10.71SLFGDD128 pKa = 4.45LFNSTVTLIDD138 pKa = 3.58NSDD141 pKa = 3.35GVPRR145 pKa = 11.84CGNCHH150 pKa = 5.93WEE152 pKa = 3.82AHH154 pKa = 6.1GPVCQHH160 pKa = 6.39CGTRR164 pKa = 11.84FRR166 pKa = 11.84VNPHH170 pKa = 5.02SRR172 pKa = 11.84FGGGYY177 pKa = 10.33DD178 pKa = 4.7DD179 pKa = 6.67DD180 pKa = 6.58DD181 pKa = 7.03DD182 pKa = 4.9EE183 pKa = 7.0DD184 pKa = 4.23EE185 pKa = 5.58EE186 pKa = 5.02SFQDD190 pKa = 4.35VIAGYY195 pKa = 9.95HH196 pKa = 6.18RR197 pKa = 11.84EE198 pKa = 3.85LDD200 pKa = 3.77TYY202 pKa = 11.3DD203 pKa = 4.37SEE205 pKa = 6.08DD206 pKa = 3.47SFIDD210 pKa = 3.14SRR212 pKa = 11.84TAHH215 pKa = 7.29EE216 pKa = 4.86IPVDD220 pKa = 3.63MSDD223 pKa = 3.88EE224 pKa = 4.64DD225 pKa = 4.91DD226 pKa = 4.56VDD228 pKa = 5.24DD229 pKa = 5.35EE230 pKa = 4.99GDD232 pKa = 3.57NEE234 pKa = 4.94RR235 pKa = 11.84GDD237 pKa = 4.6DD238 pKa = 4.61SDD240 pKa = 5.62DD241 pKa = 4.21FSDD244 pKa = 4.12NYY246 pKa = 10.76EE247 pKa = 4.13GDD249 pKa = 3.73SADD252 pKa = 3.46GDD254 pKa = 4.26RR255 pKa = 11.84NNSLLSADD263 pKa = 4.29DD264 pKa = 4.92DD265 pKa = 4.09YY266 pKa = 12.1HH267 pKa = 7.37EE268 pKa = 5.22LVLNGHH274 pKa = 7.08PADD277 pKa = 3.52EE278 pKa = 4.17WHH280 pKa = 6.54GFEE283 pKa = 4.68SASSSMIDD291 pKa = 2.82RR292 pKa = 11.84ALSNDD297 pKa = 2.84SDD299 pKa = 4.4YY300 pKa = 11.21QRR302 pKa = 11.84HH303 pKa = 5.09IHH305 pKa = 7.5DD306 pKa = 5.8DD307 pKa = 3.53NDD309 pKa = 4.29DD310 pKa = 4.14DD311 pKa = 6.13DD312 pKa = 5.52EE313 pKa = 4.62EE314 pKa = 4.96SHH316 pKa = 5.35TQEE319 pKa = 5.27IEE321 pKa = 3.75DD322 pKa = 3.89DD323 pKa = 3.75HH324 pKa = 7.47PEE326 pKa = 4.24RR327 pKa = 11.84YY328 pKa = 9.38HH329 pKa = 6.91LSSPSSEE336 pKa = 4.64NIIQLSGNDD345 pKa = 3.45DD346 pKa = 3.45DD347 pKa = 5.98SYY349 pKa = 11.8YY350 pKa = 11.01DD351 pKa = 3.61SEE353 pKa = 4.64DD354 pKa = 3.25VRR356 pKa = 11.84EE357 pKa = 4.52ALDD360 pKa = 4.73DD361 pKa = 4.44LDD363 pKa = 4.28NVEE366 pKa = 5.65ALLSYY371 pKa = 11.28DD372 pKa = 3.79NLIEE376 pKa = 4.83DD377 pKa = 3.83VSDD380 pKa = 4.94DD381 pKa = 3.92GLEE384 pKa = 4.3DD385 pKa = 5.04GYY387 pKa = 11.39DD388 pKa = 3.5SSGMVHH394 pKa = 7.1ALEE397 pKa = 5.69DD398 pKa = 5.28DD399 pKa = 3.8INLSDD404 pKa = 5.32DD405 pKa = 3.71YY406 pKa = 12.06
MM1 pKa = 8.11DD2 pKa = 5.97ANLEE6 pKa = 4.05NTKK9 pKa = 8.51MWEE12 pKa = 4.35SVDD15 pKa = 4.21HH16 pKa = 6.71SLTSNLLSRR25 pKa = 11.84ITSTVEE31 pKa = 3.46CTICSEE37 pKa = 3.86IMAVPVTAEE46 pKa = 4.35CGHH49 pKa = 5.91SFCYY53 pKa = 10.36DD54 pKa = 4.35CIYY57 pKa = 10.84QWFSNKK63 pKa = 8.75VNCPTCRR70 pKa = 11.84HH71 pKa = 5.95EE72 pKa = 5.0IEE74 pKa = 4.73NKK76 pKa = 7.93PTLNLQLKK84 pKa = 7.16EE85 pKa = 3.8ICKK88 pKa = 10.25NLLDD92 pKa = 4.43MIIDD96 pKa = 3.67SRR98 pKa = 11.84LEE100 pKa = 3.64NHH102 pKa = 6.46EE103 pKa = 3.84VLKK106 pKa = 10.49NHH108 pKa = 6.66KK109 pKa = 10.24LEE111 pKa = 4.6SEE113 pKa = 4.03NNYY116 pKa = 10.32NLDD119 pKa = 3.33VRR121 pKa = 11.84NKK123 pKa = 10.71SLFGDD128 pKa = 4.45LFNSTVTLIDD138 pKa = 3.58NSDD141 pKa = 3.35GVPRR145 pKa = 11.84CGNCHH150 pKa = 5.93WEE152 pKa = 3.82AHH154 pKa = 6.1GPVCQHH160 pKa = 6.39CGTRR164 pKa = 11.84FRR166 pKa = 11.84VNPHH170 pKa = 5.02SRR172 pKa = 11.84FGGGYY177 pKa = 10.33DD178 pKa = 4.7DD179 pKa = 6.67DD180 pKa = 6.58DD181 pKa = 7.03DD182 pKa = 4.9EE183 pKa = 7.0DD184 pKa = 4.23EE185 pKa = 5.58EE186 pKa = 5.02SFQDD190 pKa = 4.35VIAGYY195 pKa = 9.95HH196 pKa = 6.18RR197 pKa = 11.84EE198 pKa = 3.85LDD200 pKa = 3.77TYY202 pKa = 11.3DD203 pKa = 4.37SEE205 pKa = 6.08DD206 pKa = 3.47SFIDD210 pKa = 3.14SRR212 pKa = 11.84TAHH215 pKa = 7.29EE216 pKa = 4.86IPVDD220 pKa = 3.63MSDD223 pKa = 3.88EE224 pKa = 4.64DD225 pKa = 4.91DD226 pKa = 4.56VDD228 pKa = 5.24DD229 pKa = 5.35EE230 pKa = 4.99GDD232 pKa = 3.57NEE234 pKa = 4.94RR235 pKa = 11.84GDD237 pKa = 4.6DD238 pKa = 4.61SDD240 pKa = 5.62DD241 pKa = 4.21FSDD244 pKa = 4.12NYY246 pKa = 10.76EE247 pKa = 4.13GDD249 pKa = 3.73SADD252 pKa = 3.46GDD254 pKa = 4.26RR255 pKa = 11.84NNSLLSADD263 pKa = 4.29DD264 pKa = 4.92DD265 pKa = 4.09YY266 pKa = 12.1HH267 pKa = 7.37EE268 pKa = 5.22LVLNGHH274 pKa = 7.08PADD277 pKa = 3.52EE278 pKa = 4.17WHH280 pKa = 6.54GFEE283 pKa = 4.68SASSSMIDD291 pKa = 2.82RR292 pKa = 11.84ALSNDD297 pKa = 2.84SDD299 pKa = 4.4YY300 pKa = 11.21QRR302 pKa = 11.84HH303 pKa = 5.09IHH305 pKa = 7.5DD306 pKa = 5.8DD307 pKa = 3.53NDD309 pKa = 4.29DD310 pKa = 4.14DD311 pKa = 6.13DD312 pKa = 5.52EE313 pKa = 4.62EE314 pKa = 4.96SHH316 pKa = 5.35TQEE319 pKa = 5.27IEE321 pKa = 3.75DD322 pKa = 3.89DD323 pKa = 3.75HH324 pKa = 7.47PEE326 pKa = 4.24RR327 pKa = 11.84YY328 pKa = 9.38HH329 pKa = 6.91LSSPSSEE336 pKa = 4.64NIIQLSGNDD345 pKa = 3.45DD346 pKa = 3.45DD347 pKa = 5.98SYY349 pKa = 11.8YY350 pKa = 11.01DD351 pKa = 3.61SEE353 pKa = 4.64DD354 pKa = 3.25VRR356 pKa = 11.84EE357 pKa = 4.52ALDD360 pKa = 4.73DD361 pKa = 4.44LDD363 pKa = 4.28NVEE366 pKa = 5.65ALLSYY371 pKa = 11.28DD372 pKa = 3.79NLIEE376 pKa = 4.83DD377 pKa = 3.83VSDD380 pKa = 4.94DD381 pKa = 3.92GLEE384 pKa = 4.3DD385 pKa = 5.04GYY387 pKa = 11.39DD388 pKa = 3.5SSGMVHH394 pKa = 7.1ALEE397 pKa = 5.69DD398 pKa = 5.28DD399 pKa = 3.8INLSDD404 pKa = 5.32DD405 pKa = 3.71YY406 pKa = 12.06
Molecular weight: 46.08 kDa
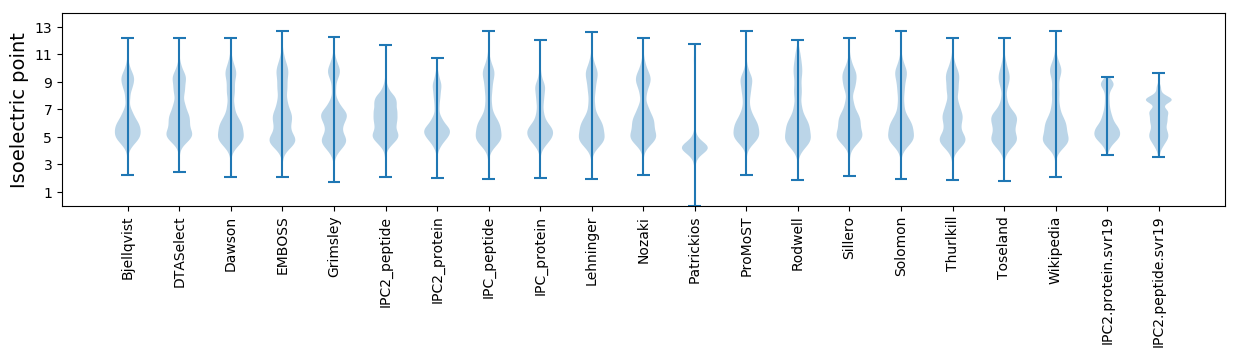
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A367XLL5|A0A367XLL5_9ASCO Vacuolar protein sorting-associated protein 33 OS=Candida viswanathii OX=5486 GN=VPS33_1 PE=3 SV=1
MM1 pKa = 7.74TIRR4 pKa = 11.84EE5 pKa = 4.2KK6 pKa = 10.8RR7 pKa = 11.84SNCSVFVPSLEE18 pKa = 4.56TKK20 pKa = 10.43CLLYY24 pKa = 9.78LTQSLCNPGGKK35 pKa = 8.89IGLFNVFANHH45 pKa = 6.56RR46 pKa = 11.84SMARR50 pKa = 11.84SNSLFGKK57 pKa = 9.11VRR59 pKa = 11.84WRR61 pKa = 11.84NGG63 pKa = 2.83
MM1 pKa = 7.74TIRR4 pKa = 11.84EE5 pKa = 4.2KK6 pKa = 10.8RR7 pKa = 11.84SNCSVFVPSLEE18 pKa = 4.56TKK20 pKa = 10.43CLLYY24 pKa = 9.78LTQSLCNPGGKK35 pKa = 8.89IGLFNVFANHH45 pKa = 6.56RR46 pKa = 11.84SMARR50 pKa = 11.84SNSLFGKK57 pKa = 9.11VRR59 pKa = 11.84WRR61 pKa = 11.84NGG63 pKa = 2.83
Molecular weight: 7.14 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5109780 |
25 |
4929 |
483.1 |
54.37 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.977 ± 0.026 | 1.071 ± 0.008 |
6.057 ± 0.022 | 6.768 ± 0.033 |
4.386 ± 0.018 | 5.434 ± 0.028 |
2.145 ± 0.012 | 6.274 ± 0.022 |
6.821 ± 0.031 | 9.508 ± 0.033 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.848 ± 0.01 | 5.449 ± 0.022 |
4.892 ± 0.028 | 4.179 ± 0.026 |
4.174 ± 0.018 | 8.297 ± 0.037 |
6.081 ± 0.042 | 6.097 ± 0.025 |
1.016 ± 0.008 | 3.527 ± 0.015 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
