
Capybara microvirus Cap3_SP_472
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 5.43
Get precalculated fractions of proteins
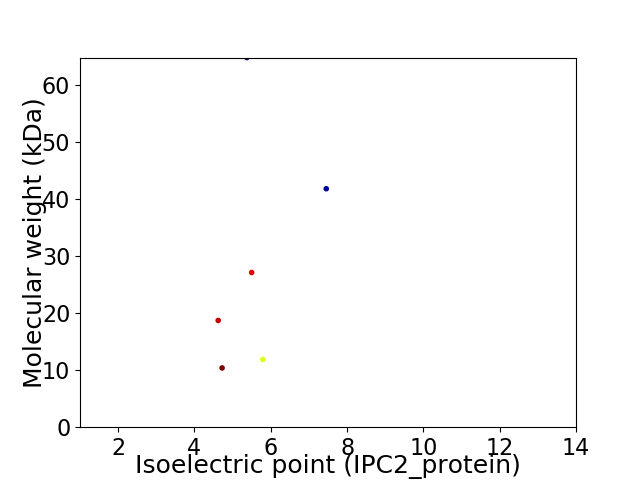
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
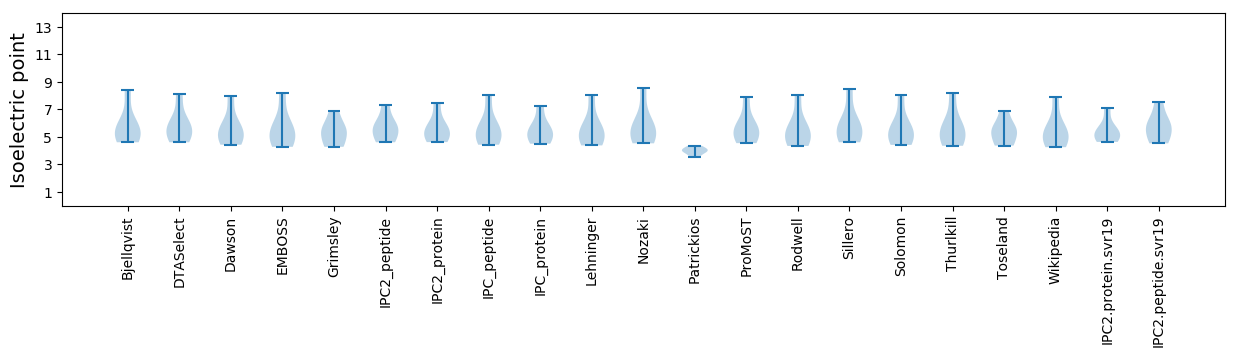
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4P8W8Q4|A0A4P8W8Q4_9VIRU Uncharacterized protein OS=Capybara microvirus Cap3_SP_472 OX=2585466 PE=4 SV=1
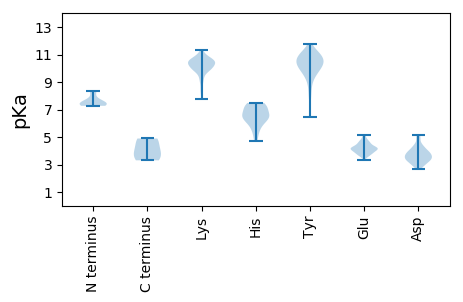
MM1 pKa = 7.61NEE3 pKa = 3.58TLLYY7 pKa = 7.65RR8 pKa = 11.84TPYY11 pKa = 10.05DD12 pKa = 3.3GKK14 pKa = 9.11TSRR17 pKa = 11.84IPTDD21 pKa = 4.33PGSEE25 pKa = 3.67WANEE29 pKa = 3.91YY30 pKa = 10.38EE31 pKa = 4.07LQIVNKK37 pKa = 10.23KK38 pKa = 9.91KK39 pKa = 10.49DD40 pKa = 3.37LVYY43 pKa = 10.24TGRR46 pKa = 11.84HH47 pKa = 4.7NLYY50 pKa = 10.87EE51 pKa = 4.49EE52 pKa = 3.69IQKK55 pKa = 10.65YY56 pKa = 9.45KK57 pKa = 10.52EE58 pKa = 3.68EE59 pKa = 4.48CEE61 pKa = 3.75IEE63 pKa = 4.71NILKK67 pKa = 10.54RR68 pKa = 11.84MSTGDD73 pKa = 3.38YY74 pKa = 11.31SMLGEE79 pKa = 4.4EE80 pKa = 4.84GNYY83 pKa = 9.55IDD85 pKa = 5.14TEE87 pKa = 4.3GMPDD91 pKa = 3.72NLMDD95 pKa = 4.22AQNSILKK102 pKa = 9.29LQNEE106 pKa = 4.35WLKK109 pKa = 11.32LPVEE113 pKa = 4.05IRR115 pKa = 11.84KK116 pKa = 9.78KK117 pKa = 10.58FDD119 pKa = 3.16NSLEE123 pKa = 4.11AFVEE127 pKa = 4.54EE128 pKa = 4.64SGSEE132 pKa = 3.61EE133 pKa = 3.99WLNKK137 pKa = 10.12LGYY140 pKa = 6.72EE141 pKa = 4.27TKK143 pKa = 10.38KK144 pKa = 10.81EE145 pKa = 3.93EE146 pKa = 4.22TMKK149 pKa = 9.55QTSTEE154 pKa = 3.95TGEE157 pKa = 4.2TNEE160 pKa = 4.07
MM1 pKa = 7.61NEE3 pKa = 3.58TLLYY7 pKa = 7.65RR8 pKa = 11.84TPYY11 pKa = 10.05DD12 pKa = 3.3GKK14 pKa = 9.11TSRR17 pKa = 11.84IPTDD21 pKa = 4.33PGSEE25 pKa = 3.67WANEE29 pKa = 3.91YY30 pKa = 10.38EE31 pKa = 4.07LQIVNKK37 pKa = 10.23KK38 pKa = 9.91KK39 pKa = 10.49DD40 pKa = 3.37LVYY43 pKa = 10.24TGRR46 pKa = 11.84HH47 pKa = 4.7NLYY50 pKa = 10.87EE51 pKa = 4.49EE52 pKa = 3.69IQKK55 pKa = 10.65YY56 pKa = 9.45KK57 pKa = 10.52EE58 pKa = 3.68EE59 pKa = 4.48CEE61 pKa = 3.75IEE63 pKa = 4.71NILKK67 pKa = 10.54RR68 pKa = 11.84MSTGDD73 pKa = 3.38YY74 pKa = 11.31SMLGEE79 pKa = 4.4EE80 pKa = 4.84GNYY83 pKa = 9.55IDD85 pKa = 5.14TEE87 pKa = 4.3GMPDD91 pKa = 3.72NLMDD95 pKa = 4.22AQNSILKK102 pKa = 9.29LQNEE106 pKa = 4.35WLKK109 pKa = 11.32LPVEE113 pKa = 4.05IRR115 pKa = 11.84KK116 pKa = 9.78KK117 pKa = 10.58FDD119 pKa = 3.16NSLEE123 pKa = 4.11AFVEE127 pKa = 4.54EE128 pKa = 4.64SGSEE132 pKa = 3.61EE133 pKa = 3.99WLNKK137 pKa = 10.12LGYY140 pKa = 6.72EE141 pKa = 4.27TKK143 pKa = 10.38KK144 pKa = 10.81EE145 pKa = 3.93EE146 pKa = 4.22TMKK149 pKa = 9.55QTSTEE154 pKa = 3.95TGEE157 pKa = 4.2TNEE160 pKa = 4.07
Molecular weight: 18.72 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4P8W624|A0A4P8W624_9VIRU Nonstructural protein OS=Capybara microvirus Cap3_SP_472 OX=2585466 PE=4 SV=1
MM1 pKa = 7.41GCKK4 pKa = 9.4KK5 pKa = 10.31PKK7 pKa = 10.28IIVQDD12 pKa = 3.74LNKK15 pKa = 9.82WKK17 pKa = 8.97TAADD21 pKa = 3.29GHH23 pKa = 6.42KK24 pKa = 10.17YY25 pKa = 10.02HH26 pKa = 7.25PAEE29 pKa = 4.17CNYY32 pKa = 10.76ASEE35 pKa = 4.54EE36 pKa = 4.59DD37 pKa = 3.41INTYY41 pKa = 10.35KK42 pKa = 10.71DD43 pKa = 3.47EE44 pKa = 4.68KK45 pKa = 10.53IKK47 pKa = 10.75IIGTHH52 pKa = 6.33KK53 pKa = 10.7YY54 pKa = 10.62SLVPCGQCLGCRR66 pKa = 11.84LQRR69 pKa = 11.84SAEE72 pKa = 3.89WADD75 pKa = 3.57RR76 pKa = 11.84CTLEE80 pKa = 4.2MEE82 pKa = 4.79EE83 pKa = 5.17NPEE86 pKa = 4.03QCWFLTLTYY95 pKa = 10.6NDD97 pKa = 4.03EE98 pKa = 4.15NLPYY102 pKa = 9.66IDD104 pKa = 5.11EE105 pKa = 4.39FTTKK109 pKa = 10.44KK110 pKa = 10.73GITYY114 pKa = 9.62TDD116 pKa = 3.76TEE118 pKa = 4.66GTWNGCLQKK127 pKa = 10.68KK128 pKa = 10.03DD129 pKa = 3.24IQDD132 pKa = 3.2FLKK135 pKa = 10.32RR136 pKa = 11.84LRR138 pKa = 11.84RR139 pKa = 11.84RR140 pKa = 11.84WEE142 pKa = 3.96YY143 pKa = 10.84KK144 pKa = 10.29YY145 pKa = 10.27SAHH148 pKa = 7.27NIRR151 pKa = 11.84TFYY154 pKa = 10.45CGEE157 pKa = 4.06YY158 pKa = 10.48GPQTEE163 pKa = 4.19RR164 pKa = 11.84PHH166 pKa = 7.48YY167 pKa = 10.91HH168 pKa = 6.4MICFNLPITPDD179 pKa = 2.71MMTIQKK185 pKa = 10.16LNWEE189 pKa = 3.96KK190 pKa = 11.08DD191 pKa = 3.01ILYY194 pKa = 9.96KK195 pKa = 10.41CDD197 pKa = 4.07EE198 pKa = 4.66IEE200 pKa = 4.25QIWGKK205 pKa = 9.94GFVVIEE211 pKa = 4.06EE212 pKa = 4.97CNWQACNYY220 pKa = 6.53VARR223 pKa = 11.84YY224 pKa = 6.45ITKK227 pKa = 9.9KK228 pKa = 8.41QTGKK232 pKa = 10.52LADD235 pKa = 3.61EE236 pKa = 5.03YY237 pKa = 10.88YY238 pKa = 10.3AQKK241 pKa = 10.79GQIPPFVQASLKK253 pKa = 9.94PAIGKK258 pKa = 9.52NYY260 pKa = 8.41YY261 pKa = 9.46QEE263 pKa = 4.33NKK265 pKa = 10.22SKK267 pKa = 10.26IYY269 pKa = 10.63EE270 pKa = 4.08NDD272 pKa = 3.26NIIITGKK279 pKa = 10.81GKK281 pKa = 9.44VLKK284 pKa = 9.94IKK286 pKa = 9.9PPRR289 pKa = 11.84YY290 pKa = 9.0FDD292 pKa = 3.42KK293 pKa = 11.02LYY295 pKa = 11.13DD296 pKa = 3.92LDD298 pKa = 4.41YY299 pKa = 10.52PSDD302 pKa = 3.38MLRR305 pKa = 11.84IKK307 pKa = 10.5KK308 pKa = 9.86LRR310 pKa = 11.84EE311 pKa = 3.5RR312 pKa = 11.84KK313 pKa = 9.85AMNGLLLKK321 pKa = 8.72TQNTSLDD328 pKa = 3.55YY329 pKa = 11.38LEE331 pKa = 4.02QLKK334 pKa = 10.36IEE336 pKa = 4.22EE337 pKa = 5.04DD338 pKa = 4.1YY339 pKa = 10.58LTEE342 pKa = 3.87RR343 pKa = 11.84TTTLKK348 pKa = 9.76RR349 pKa = 11.84TNAEE353 pKa = 3.81AA354 pKa = 3.93
MM1 pKa = 7.41GCKK4 pKa = 9.4KK5 pKa = 10.31PKK7 pKa = 10.28IIVQDD12 pKa = 3.74LNKK15 pKa = 9.82WKK17 pKa = 8.97TAADD21 pKa = 3.29GHH23 pKa = 6.42KK24 pKa = 10.17YY25 pKa = 10.02HH26 pKa = 7.25PAEE29 pKa = 4.17CNYY32 pKa = 10.76ASEE35 pKa = 4.54EE36 pKa = 4.59DD37 pKa = 3.41INTYY41 pKa = 10.35KK42 pKa = 10.71DD43 pKa = 3.47EE44 pKa = 4.68KK45 pKa = 10.53IKK47 pKa = 10.75IIGTHH52 pKa = 6.33KK53 pKa = 10.7YY54 pKa = 10.62SLVPCGQCLGCRR66 pKa = 11.84LQRR69 pKa = 11.84SAEE72 pKa = 3.89WADD75 pKa = 3.57RR76 pKa = 11.84CTLEE80 pKa = 4.2MEE82 pKa = 4.79EE83 pKa = 5.17NPEE86 pKa = 4.03QCWFLTLTYY95 pKa = 10.6NDD97 pKa = 4.03EE98 pKa = 4.15NLPYY102 pKa = 9.66IDD104 pKa = 5.11EE105 pKa = 4.39FTTKK109 pKa = 10.44KK110 pKa = 10.73GITYY114 pKa = 9.62TDD116 pKa = 3.76TEE118 pKa = 4.66GTWNGCLQKK127 pKa = 10.68KK128 pKa = 10.03DD129 pKa = 3.24IQDD132 pKa = 3.2FLKK135 pKa = 10.32RR136 pKa = 11.84LRR138 pKa = 11.84RR139 pKa = 11.84RR140 pKa = 11.84WEE142 pKa = 3.96YY143 pKa = 10.84KK144 pKa = 10.29YY145 pKa = 10.27SAHH148 pKa = 7.27NIRR151 pKa = 11.84TFYY154 pKa = 10.45CGEE157 pKa = 4.06YY158 pKa = 10.48GPQTEE163 pKa = 4.19RR164 pKa = 11.84PHH166 pKa = 7.48YY167 pKa = 10.91HH168 pKa = 6.4MICFNLPITPDD179 pKa = 2.71MMTIQKK185 pKa = 10.16LNWEE189 pKa = 3.96KK190 pKa = 11.08DD191 pKa = 3.01ILYY194 pKa = 9.96KK195 pKa = 10.41CDD197 pKa = 4.07EE198 pKa = 4.66IEE200 pKa = 4.25QIWGKK205 pKa = 9.94GFVVIEE211 pKa = 4.06EE212 pKa = 4.97CNWQACNYY220 pKa = 6.53VARR223 pKa = 11.84YY224 pKa = 6.45ITKK227 pKa = 9.9KK228 pKa = 8.41QTGKK232 pKa = 10.52LADD235 pKa = 3.61EE236 pKa = 5.03YY237 pKa = 10.88YY238 pKa = 10.3AQKK241 pKa = 10.79GQIPPFVQASLKK253 pKa = 9.94PAIGKK258 pKa = 9.52NYY260 pKa = 8.41YY261 pKa = 9.46QEE263 pKa = 4.33NKK265 pKa = 10.22SKK267 pKa = 10.26IYY269 pKa = 10.63EE270 pKa = 4.08NDD272 pKa = 3.26NIIITGKK279 pKa = 10.81GKK281 pKa = 9.44VLKK284 pKa = 9.94IKK286 pKa = 9.9PPRR289 pKa = 11.84YY290 pKa = 9.0FDD292 pKa = 3.42KK293 pKa = 11.02LYY295 pKa = 11.13DD296 pKa = 3.92LDD298 pKa = 4.41YY299 pKa = 10.52PSDD302 pKa = 3.38MLRR305 pKa = 11.84IKK307 pKa = 10.5KK308 pKa = 9.86LRR310 pKa = 11.84EE311 pKa = 3.5RR312 pKa = 11.84KK313 pKa = 9.85AMNGLLLKK321 pKa = 8.72TQNTSLDD328 pKa = 3.55YY329 pKa = 11.38LEE331 pKa = 4.02QLKK334 pKa = 10.36IEE336 pKa = 4.22EE337 pKa = 5.04DD338 pKa = 4.1YY339 pKa = 10.58LTEE342 pKa = 3.87RR343 pKa = 11.84TTTLKK348 pKa = 9.76RR349 pKa = 11.84TNAEE353 pKa = 3.81AA354 pKa = 3.93
Molecular weight: 41.83 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1536 |
88 |
571 |
256.0 |
29.13 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.641 ± 1.872 | 1.432 ± 0.728 |
4.883 ± 0.65 | 8.398 ± 1.773 |
2.539 ± 0.548 | 6.641 ± 1.057 |
1.628 ± 0.396 | 7.096 ± 1.281 |
7.422 ± 1.673 | 6.51 ± 0.837 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.734 ± 0.364 | 6.576 ± 0.911 |
3.711 ± 1.087 | 4.557 ± 0.501 |
3.776 ± 0.345 | 7.747 ± 3.347 |
6.706 ± 0.868 | 3.19 ± 1.06 |
2.279 ± 0.225 | 5.534 ± 0.674 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
