
Roseomonas stagni DSM 19981
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodospirillales; Acetobacteraceae; Roseomonas; Roseomonas stagni
Average proteome isoelectric point is 7.01
Get precalculated fractions of proteins
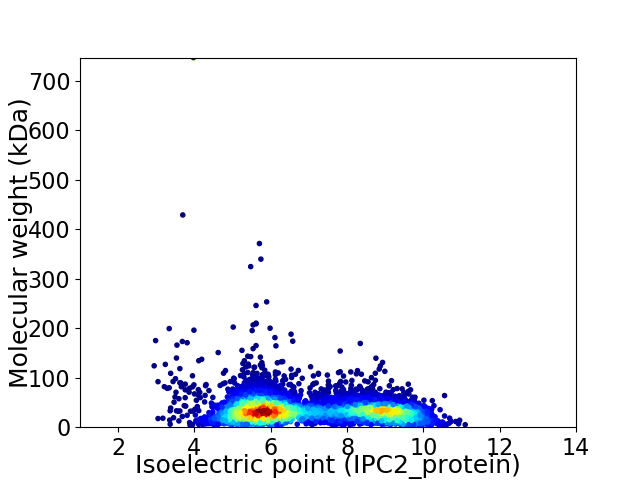
Virtual 2D-PAGE plot for 5902 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1I3XKI8|A0A1I3XKI8_9PROT Uncharacterized protein OS=Roseomonas stagni DSM 19981 OX=1123062 GN=SAMN02745775_101406 PE=4 SV=1
MM1 pKa = 7.21SQTLSGDD8 pKa = 3.29NVTDD12 pKa = 5.64SIFTWTFTANSGDD25 pKa = 3.47SWGGTLVDD33 pKa = 4.27DD34 pKa = 4.41GTRR37 pKa = 11.84YY38 pKa = 10.06VVGSILNTATGRR50 pKa = 11.84YY51 pKa = 7.22TIVAAAPQGVDD62 pKa = 3.4LSAFGMNEE70 pKa = 3.1GWIAVSWYY78 pKa = 9.7RR79 pKa = 11.84DD80 pKa = 3.13ASGVFMVTRR89 pKa = 11.84NGAGTASGTAGLGSEE104 pKa = 5.65VDD106 pKa = 3.74AAWNGTAWDD115 pKa = 3.9SFGLGGSDD123 pKa = 3.75QADD126 pKa = 3.87PGDD129 pKa = 4.0LADD132 pKa = 6.36SLFTWTFTADD142 pKa = 3.59SGDD145 pKa = 3.45IMQGTLLDD153 pKa = 4.18DD154 pKa = 3.68TRR156 pKa = 11.84NWDD159 pKa = 3.46PGDD162 pKa = 3.5TRR164 pKa = 11.84RR165 pKa = 11.84TAFGTYY171 pKa = 9.96RR172 pKa = 11.84IDD174 pKa = 3.41TEE176 pKa = 3.97IPYY179 pKa = 10.8GRR181 pKa = 11.84DD182 pKa = 2.89LGSAGVEE189 pKa = 3.99GTITIVSYY197 pKa = 11.21TDD199 pKa = 3.76FFADD203 pKa = 3.04IQFTLEE209 pKa = 3.96TGASGPAGYY218 pKa = 10.6GGFGSEE224 pKa = 4.96WDD226 pKa = 3.98RR227 pKa = 11.84AWTGTAWMPVGQGGALQADD246 pKa = 4.68RR247 pKa = 11.84LPDD250 pKa = 3.41RR251 pKa = 11.84VFSWRR256 pKa = 11.84FTADD260 pKa = 2.85NGDD263 pKa = 3.64QYY265 pKa = 11.05VGSMVGHH272 pKa = 6.41SSALTTGDD280 pKa = 4.53RR281 pKa = 11.84IEE283 pKa = 4.6TDD285 pKa = 2.77HH286 pKa = 6.11GHH288 pKa = 6.51YY289 pKa = 10.59DD290 pKa = 3.25ILQEE294 pKa = 4.06NAYY297 pKa = 9.97SGPASAQGMVWVLRR311 pKa = 11.84YY312 pKa = 9.85YY313 pKa = 10.6DD314 pKa = 4.01ASLDD318 pKa = 3.54TWMTTYY324 pKa = 10.6QNSVATTASGTRR336 pKa = 11.84GLGSEE341 pKa = 4.83YY342 pKa = 10.7DD343 pKa = 3.98YY344 pKa = 11.89AWDD347 pKa = 3.67GDD349 pKa = 3.55EE350 pKa = 4.58WDD352 pKa = 4.55DD353 pKa = 4.0FGLGGVHH360 pKa = 7.0LASIEE365 pKa = 4.07RR366 pKa = 11.84NNLFGWTFTANSGDD380 pKa = 4.67RR381 pKa = 11.84YY382 pKa = 8.85TGWLIAADD390 pKa = 3.91DD391 pKa = 4.63AYY393 pKa = 10.95AAGDD397 pKa = 3.98TVAGAQGRR405 pKa = 11.84YY406 pKa = 8.81QITYY410 pKa = 8.17EE411 pKa = 4.29GTWGGAAGAGTIYY424 pKa = 7.44TTSYY428 pKa = 10.95SDD430 pKa = 3.25AGSGRR435 pKa = 11.84SFDD438 pKa = 3.52TYY440 pKa = 10.49RR441 pKa = 11.84WGTVGGQPSGRR452 pKa = 11.84AGFGSEE458 pKa = 5.35YY459 pKa = 10.64DD460 pKa = 4.06YY461 pKa = 11.3IWNGSEE467 pKa = 3.48WDD469 pKa = 3.74DD470 pKa = 4.13FGVGGVHH477 pKa = 5.76QANYY481 pKa = 9.17TINTLHH487 pKa = 7.13AFTFTANSGDD497 pKa = 4.63RR498 pKa = 11.84YY499 pKa = 8.92TGWLVEE505 pKa = 4.25DD506 pKa = 3.66SARR509 pKa = 11.84YY510 pKa = 9.1AVGNMIATSQGSYY523 pKa = 10.15RR524 pKa = 11.84ITWEE528 pKa = 4.1GDD530 pKa = 2.49WGGTQGRR537 pKa = 11.84GSIWTDD543 pKa = 2.91SYY545 pKa = 11.71FDD547 pKa = 3.74AASGRR552 pKa = 11.84SFGTYY557 pKa = 7.03TWTAGGGRR565 pKa = 11.84PSGQAGFGSEE575 pKa = 4.42YY576 pKa = 10.75DD577 pKa = 4.14YY578 pKa = 11.29IWTGTEE584 pKa = 3.28WDD586 pKa = 3.9DD587 pKa = 4.11FGVGGVHH594 pKa = 6.56QSTFTINTLHH604 pKa = 7.04AFTFTASSGDD614 pKa = 3.87RR615 pKa = 11.84YY616 pKa = 10.78SGWLIEE622 pKa = 4.54DD623 pKa = 3.46STRR626 pKa = 11.84YY627 pKa = 9.91AVGDD631 pKa = 4.22TITTSQGSYY640 pKa = 10.36RR641 pKa = 11.84ITFEE645 pKa = 4.15GDD647 pKa = 2.26WGGTQGRR654 pKa = 11.84GSIWTDD660 pKa = 2.87SYY662 pKa = 11.77FDD664 pKa = 3.88AGTGRR669 pKa = 11.84SFATYY674 pKa = 9.57FWGTLGAQPSGRR686 pKa = 11.84SGFGSEE692 pKa = 5.29YY693 pKa = 10.75DD694 pKa = 4.09YY695 pKa = 11.4IWDD698 pKa = 3.64GDD700 pKa = 3.47EE701 pKa = 3.88WDD703 pKa = 4.25DD704 pKa = 4.18FGVGGIHH711 pKa = 5.96QANVTVNTLFGWTFTANSGDD731 pKa = 4.67RR732 pKa = 11.84YY733 pKa = 8.87TGWLIQDD740 pKa = 3.52DD741 pKa = 3.64TRR743 pKa = 11.84YY744 pKa = 10.59AVGDD748 pKa = 4.27TIATSQGSYY757 pKa = 10.35RR758 pKa = 11.84ITHH761 pKa = 6.29EE762 pKa = 4.54GPWGGTAGKK771 pKa = 8.11NTIWTTSYY779 pKa = 11.08FDD781 pKa = 3.8AGSNQTFEE789 pKa = 4.39TYY791 pKa = 9.3FWNTIGGQPSGRR803 pKa = 11.84GGFGSEE809 pKa = 4.17YY810 pKa = 10.52DD811 pKa = 3.97YY812 pKa = 11.88VWDD815 pKa = 3.82GRR817 pKa = 11.84EE818 pKa = 3.64WDD820 pKa = 3.86DD821 pKa = 4.11FGVGGVHH828 pKa = 5.1QVRR831 pKa = 11.84VEE833 pKa = 4.01RR834 pKa = 11.84AVMVSWIFRR843 pKa = 11.84ATSGDD848 pKa = 3.88AYY850 pKa = 9.97TGLLLEE856 pKa = 4.98DD857 pKa = 4.49AEE859 pKa = 4.95TYY861 pKa = 11.11APGDD865 pKa = 3.65TIVRR869 pKa = 11.84PNGTYY874 pKa = 10.09TIQSEE879 pKa = 4.46TDD881 pKa = 3.26TNRR884 pKa = 11.84TDD886 pKa = 3.52YY887 pKa = 11.49GIGTIWTAAYY897 pKa = 10.02YY898 pKa = 10.29DD899 pKa = 4.15AGSATWLTTHH909 pKa = 6.82YY910 pKa = 10.68YY911 pKa = 10.69GFYY914 pKa = 10.66DD915 pKa = 4.18GLPSGRR921 pKa = 11.84SGFGSEE927 pKa = 5.84YY928 pKa = 11.1DD929 pKa = 3.86FAWDD933 pKa = 3.24GDD935 pKa = 3.55EE936 pKa = 3.99WDD938 pKa = 4.37DD939 pKa = 4.25FGVAGLHH946 pKa = 5.23QADD949 pKa = 4.29VEE951 pKa = 4.31RR952 pKa = 11.84ASAPEE957 pKa = 3.72VLGG960 pKa = 3.95
MM1 pKa = 7.21SQTLSGDD8 pKa = 3.29NVTDD12 pKa = 5.64SIFTWTFTANSGDD25 pKa = 3.47SWGGTLVDD33 pKa = 4.27DD34 pKa = 4.41GTRR37 pKa = 11.84YY38 pKa = 10.06VVGSILNTATGRR50 pKa = 11.84YY51 pKa = 7.22TIVAAAPQGVDD62 pKa = 3.4LSAFGMNEE70 pKa = 3.1GWIAVSWYY78 pKa = 9.7RR79 pKa = 11.84DD80 pKa = 3.13ASGVFMVTRR89 pKa = 11.84NGAGTASGTAGLGSEE104 pKa = 5.65VDD106 pKa = 3.74AAWNGTAWDD115 pKa = 3.9SFGLGGSDD123 pKa = 3.75QADD126 pKa = 3.87PGDD129 pKa = 4.0LADD132 pKa = 6.36SLFTWTFTADD142 pKa = 3.59SGDD145 pKa = 3.45IMQGTLLDD153 pKa = 4.18DD154 pKa = 3.68TRR156 pKa = 11.84NWDD159 pKa = 3.46PGDD162 pKa = 3.5TRR164 pKa = 11.84RR165 pKa = 11.84TAFGTYY171 pKa = 9.96RR172 pKa = 11.84IDD174 pKa = 3.41TEE176 pKa = 3.97IPYY179 pKa = 10.8GRR181 pKa = 11.84DD182 pKa = 2.89LGSAGVEE189 pKa = 3.99GTITIVSYY197 pKa = 11.21TDD199 pKa = 3.76FFADD203 pKa = 3.04IQFTLEE209 pKa = 3.96TGASGPAGYY218 pKa = 10.6GGFGSEE224 pKa = 4.96WDD226 pKa = 3.98RR227 pKa = 11.84AWTGTAWMPVGQGGALQADD246 pKa = 4.68RR247 pKa = 11.84LPDD250 pKa = 3.41RR251 pKa = 11.84VFSWRR256 pKa = 11.84FTADD260 pKa = 2.85NGDD263 pKa = 3.64QYY265 pKa = 11.05VGSMVGHH272 pKa = 6.41SSALTTGDD280 pKa = 4.53RR281 pKa = 11.84IEE283 pKa = 4.6TDD285 pKa = 2.77HH286 pKa = 6.11GHH288 pKa = 6.51YY289 pKa = 10.59DD290 pKa = 3.25ILQEE294 pKa = 4.06NAYY297 pKa = 9.97SGPASAQGMVWVLRR311 pKa = 11.84YY312 pKa = 9.85YY313 pKa = 10.6DD314 pKa = 4.01ASLDD318 pKa = 3.54TWMTTYY324 pKa = 10.6QNSVATTASGTRR336 pKa = 11.84GLGSEE341 pKa = 4.83YY342 pKa = 10.7DD343 pKa = 3.98YY344 pKa = 11.89AWDD347 pKa = 3.67GDD349 pKa = 3.55EE350 pKa = 4.58WDD352 pKa = 4.55DD353 pKa = 4.0FGLGGVHH360 pKa = 7.0LASIEE365 pKa = 4.07RR366 pKa = 11.84NNLFGWTFTANSGDD380 pKa = 4.67RR381 pKa = 11.84YY382 pKa = 8.85TGWLIAADD390 pKa = 3.91DD391 pKa = 4.63AYY393 pKa = 10.95AAGDD397 pKa = 3.98TVAGAQGRR405 pKa = 11.84YY406 pKa = 8.81QITYY410 pKa = 8.17EE411 pKa = 4.29GTWGGAAGAGTIYY424 pKa = 7.44TTSYY428 pKa = 10.95SDD430 pKa = 3.25AGSGRR435 pKa = 11.84SFDD438 pKa = 3.52TYY440 pKa = 10.49RR441 pKa = 11.84WGTVGGQPSGRR452 pKa = 11.84AGFGSEE458 pKa = 5.35YY459 pKa = 10.64DD460 pKa = 4.06YY461 pKa = 11.3IWNGSEE467 pKa = 3.48WDD469 pKa = 3.74DD470 pKa = 4.13FGVGGVHH477 pKa = 5.76QANYY481 pKa = 9.17TINTLHH487 pKa = 7.13AFTFTANSGDD497 pKa = 4.63RR498 pKa = 11.84YY499 pKa = 8.92TGWLVEE505 pKa = 4.25DD506 pKa = 3.66SARR509 pKa = 11.84YY510 pKa = 9.1AVGNMIATSQGSYY523 pKa = 10.15RR524 pKa = 11.84ITWEE528 pKa = 4.1GDD530 pKa = 2.49WGGTQGRR537 pKa = 11.84GSIWTDD543 pKa = 2.91SYY545 pKa = 11.71FDD547 pKa = 3.74AASGRR552 pKa = 11.84SFGTYY557 pKa = 7.03TWTAGGGRR565 pKa = 11.84PSGQAGFGSEE575 pKa = 4.42YY576 pKa = 10.75DD577 pKa = 4.14YY578 pKa = 11.29IWTGTEE584 pKa = 3.28WDD586 pKa = 3.9DD587 pKa = 4.11FGVGGVHH594 pKa = 6.56QSTFTINTLHH604 pKa = 7.04AFTFTASSGDD614 pKa = 3.87RR615 pKa = 11.84YY616 pKa = 10.78SGWLIEE622 pKa = 4.54DD623 pKa = 3.46STRR626 pKa = 11.84YY627 pKa = 9.91AVGDD631 pKa = 4.22TITTSQGSYY640 pKa = 10.36RR641 pKa = 11.84ITFEE645 pKa = 4.15GDD647 pKa = 2.26WGGTQGRR654 pKa = 11.84GSIWTDD660 pKa = 2.87SYY662 pKa = 11.77FDD664 pKa = 3.88AGTGRR669 pKa = 11.84SFATYY674 pKa = 9.57FWGTLGAQPSGRR686 pKa = 11.84SGFGSEE692 pKa = 5.29YY693 pKa = 10.75DD694 pKa = 4.09YY695 pKa = 11.4IWDD698 pKa = 3.64GDD700 pKa = 3.47EE701 pKa = 3.88WDD703 pKa = 4.25DD704 pKa = 4.18FGVGGIHH711 pKa = 5.96QANVTVNTLFGWTFTANSGDD731 pKa = 4.67RR732 pKa = 11.84YY733 pKa = 8.87TGWLIQDD740 pKa = 3.52DD741 pKa = 3.64TRR743 pKa = 11.84YY744 pKa = 10.59AVGDD748 pKa = 4.27TIATSQGSYY757 pKa = 10.35RR758 pKa = 11.84ITHH761 pKa = 6.29EE762 pKa = 4.54GPWGGTAGKK771 pKa = 8.11NTIWTTSYY779 pKa = 11.08FDD781 pKa = 3.8AGSNQTFEE789 pKa = 4.39TYY791 pKa = 9.3FWNTIGGQPSGRR803 pKa = 11.84GGFGSEE809 pKa = 4.17YY810 pKa = 10.52DD811 pKa = 3.97YY812 pKa = 11.88VWDD815 pKa = 3.82GRR817 pKa = 11.84EE818 pKa = 3.64WDD820 pKa = 3.86DD821 pKa = 4.11FGVGGVHH828 pKa = 5.1QVRR831 pKa = 11.84VEE833 pKa = 4.01RR834 pKa = 11.84AVMVSWIFRR843 pKa = 11.84ATSGDD848 pKa = 3.88AYY850 pKa = 9.97TGLLLEE856 pKa = 4.98DD857 pKa = 4.49AEE859 pKa = 4.95TYY861 pKa = 11.11APGDD865 pKa = 3.65TIVRR869 pKa = 11.84PNGTYY874 pKa = 10.09TIQSEE879 pKa = 4.46TDD881 pKa = 3.26TNRR884 pKa = 11.84TDD886 pKa = 3.52YY887 pKa = 11.49GIGTIWTAAYY897 pKa = 10.02YY898 pKa = 10.29DD899 pKa = 4.15AGSATWLTTHH909 pKa = 6.82YY910 pKa = 10.68YY911 pKa = 10.69GFYY914 pKa = 10.66DD915 pKa = 4.18GLPSGRR921 pKa = 11.84SGFGSEE927 pKa = 5.84YY928 pKa = 11.1DD929 pKa = 3.86FAWDD933 pKa = 3.24GDD935 pKa = 3.55EE936 pKa = 3.99WDD938 pKa = 4.37DD939 pKa = 4.25FGVAGLHH946 pKa = 5.23QADD949 pKa = 4.29VEE951 pKa = 4.31RR952 pKa = 11.84ASAPEE957 pKa = 3.72VLGG960 pKa = 3.95
Molecular weight: 103.8 kDa
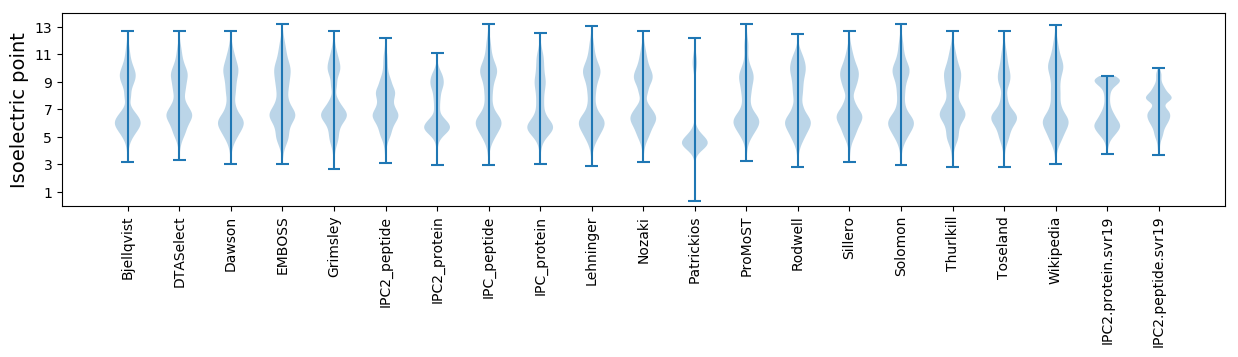
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1I4ABG7|A0A1I4ABG7_9PROT Phosphoenolpyruvate-protein phosphotransferase OS=Roseomonas stagni DSM 19981 OX=1123062 GN=SAMN02745775_103291 PE=3 SV=1
MM1 pKa = 7.88RR2 pKa = 11.84SLSRR6 pKa = 11.84LPLAATLLLIGAVPAFAQGSSGTVTSTAPVATTRR40 pKa = 11.84PAVTAPAAPTQRR52 pKa = 11.84PTQAATPANPGGSAQATAPAARR74 pKa = 11.84PATQGQQAQPATPAVPQAPRR94 pKa = 11.84ATNN97 pKa = 3.19
MM1 pKa = 7.88RR2 pKa = 11.84SLSRR6 pKa = 11.84LPLAATLLLIGAVPAFAQGSSGTVTSTAPVATTRR40 pKa = 11.84PAVTAPAAPTQRR52 pKa = 11.84PTQAATPANPGGSAQATAPAARR74 pKa = 11.84PATQGQQAQPATPAVPQAPRR94 pKa = 11.84ATNN97 pKa = 3.19
Molecular weight: 9.42 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1962004 |
8 |
7448 |
332.4 |
35.41 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
15.48 ± 0.059 | 0.784 ± 0.011 |
5.323 ± 0.038 | 5.102 ± 0.03 |
3.246 ± 0.021 | 9.502 ± 0.054 |
1.931 ± 0.017 | 4.282 ± 0.024 |
1.808 ± 0.023 | 10.685 ± 0.042 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.484 ± 0.017 | 2.034 ± 0.018 |
6.256 ± 0.038 | 2.905 ± 0.019 |
8.192 ± 0.039 | 4.324 ± 0.033 |
5.198 ± 0.038 | 7.425 ± 0.028 |
1.432 ± 0.014 | 1.608 ± 0.012 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
