
Mesocricetus auratus papillomavirus 1
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Pipapillomavirus; Pipapillomavirus 1
Average proteome isoelectric point is 6.19
Get precalculated fractions of proteins
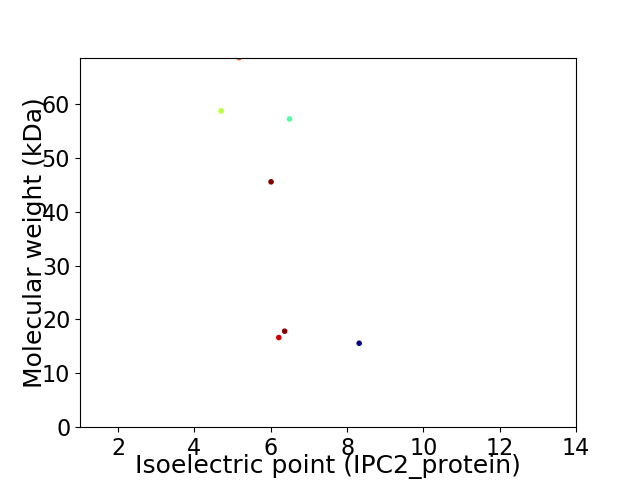
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|U6ELS6|U6ELS6_9PAPI Protein E7 OS=Mesocricetus auratus papillomavirus 1 OX=1408129 GN=E7 PE=3 SV=1
MM1 pKa = 6.87VAANRR6 pKa = 11.84ARR8 pKa = 11.84RR9 pKa = 11.84VKK11 pKa = 10.38RR12 pKa = 11.84DD13 pKa = 3.24SAEE16 pKa = 3.68NLYY19 pKa = 10.02RR20 pKa = 11.84QCQITGNCPPDD31 pKa = 3.47VVNKK35 pKa = 10.28VEE37 pKa = 4.13GTTLADD43 pKa = 3.33KK44 pKa = 9.14LTKK47 pKa = 10.05IFASILYY54 pKa = 10.19LGGLGIGTGRR64 pKa = 11.84GTGGATGYY72 pKa = 11.23GPINPGGGRR81 pKa = 11.84ITGTGTVMRR90 pKa = 11.84PGVVIEE96 pKa = 4.19PVGPGDD102 pKa = 3.56IVSVGATDD110 pKa = 3.67SSIVPLLEE118 pKa = 4.09ATPDD122 pKa = 3.28IPIEE126 pKa = 4.48GGPEE130 pKa = 3.79VPPAGPDD137 pKa = 3.19VSTVDD142 pKa = 3.03VTANVDD148 pKa = 4.11PISDD152 pKa = 3.69INVTSSTTITNPDD165 pKa = 3.32SAVIDD170 pKa = 3.93VQPAPSGPRR179 pKa = 11.84RR180 pKa = 11.84VTVSRR185 pKa = 11.84SEE187 pKa = 4.07FQNASYY193 pKa = 11.39VSVTHH198 pKa = 6.8PSQGLGEE205 pKa = 4.33SGGAVISADD214 pKa = 3.1AGGSVVSSSHH224 pKa = 5.69EE225 pKa = 3.95LDD227 pKa = 3.23VGILIGGRR235 pKa = 11.84PPGNLSEE242 pKa = 4.23FQTIEE247 pKa = 3.89LDD249 pKa = 4.73EE250 pKa = 4.4IVQTQGVDD258 pKa = 3.23DD259 pKa = 4.27FDD261 pKa = 4.56IGEE264 pKa = 4.42GAGRR268 pKa = 11.84AGPDD272 pKa = 2.79TSTPDD277 pKa = 3.23NYY279 pKa = 10.81LGRR282 pKa = 11.84ALSRR286 pKa = 11.84FRR288 pKa = 11.84EE289 pKa = 4.31IYY291 pKa = 9.64EE292 pKa = 4.33RR293 pKa = 11.84PSDD296 pKa = 3.7LYY298 pKa = 9.96TRR300 pKa = 11.84RR301 pKa = 11.84VQQVRR306 pKa = 11.84VRR308 pKa = 11.84NRR310 pKa = 11.84DD311 pKa = 3.25LFLGDD316 pKa = 3.54PGRR319 pKa = 11.84LVTYY323 pKa = 10.03EE324 pKa = 3.81FDD326 pKa = 3.52NPAFEE331 pKa = 5.13GLGEE335 pKa = 4.03DD336 pKa = 4.41TIIFHH341 pKa = 6.45QAPEE345 pKa = 4.13VQAAPDD351 pKa = 4.09EE352 pKa = 4.47DD353 pKa = 4.3FMDD356 pKa = 4.9IIKK359 pKa = 10.7LGRR362 pKa = 11.84QRR364 pKa = 11.84LAQTPGGTVRR374 pKa = 11.84ISRR377 pKa = 11.84LGQRR381 pKa = 11.84GTIRR385 pKa = 11.84TRR387 pKa = 11.84SGLQIGGKK395 pKa = 7.39VHH397 pKa = 6.87FFSDD401 pKa = 4.05LSPIATEE408 pKa = 4.38NIEE411 pKa = 4.18LSPLGEE417 pKa = 3.97ISGEE421 pKa = 3.98AMVVDD426 pKa = 4.58ALSEE430 pKa = 3.98YY431 pKa = 11.25SLISEE436 pKa = 4.63PGIEE440 pKa = 4.44PVPFMDD446 pKa = 5.44EE447 pKa = 4.32DD448 pKa = 3.97LLDD451 pKa = 3.85IQSEE455 pKa = 4.34DD456 pKa = 3.57FSGSRR461 pKa = 11.84LHH463 pKa = 7.19LVRR466 pKa = 11.84TGNRR470 pKa = 11.84FGVIYY475 pKa = 10.06EE476 pKa = 3.9PSEE479 pKa = 3.78NLGARR484 pKa = 11.84TVFPDD489 pKa = 3.15INTGDD494 pKa = 4.9FIRR497 pKa = 11.84YY498 pKa = 5.82PQSNINPADD507 pKa = 3.29ISYY510 pKa = 11.08QDD512 pKa = 5.66LIPLQPGVDD521 pKa = 3.35INVGSIYY528 pKa = 10.24PSVDD532 pKa = 3.37YY533 pKa = 10.69YY534 pKa = 11.14LHH536 pKa = 6.65PSLRR540 pKa = 11.84RR541 pKa = 11.84RR542 pKa = 11.84KK543 pKa = 9.68RR544 pKa = 11.84KK545 pKa = 8.56RR546 pKa = 11.84TLHH549 pKa = 5.67
MM1 pKa = 6.87VAANRR6 pKa = 11.84ARR8 pKa = 11.84RR9 pKa = 11.84VKK11 pKa = 10.38RR12 pKa = 11.84DD13 pKa = 3.24SAEE16 pKa = 3.68NLYY19 pKa = 10.02RR20 pKa = 11.84QCQITGNCPPDD31 pKa = 3.47VVNKK35 pKa = 10.28VEE37 pKa = 4.13GTTLADD43 pKa = 3.33KK44 pKa = 9.14LTKK47 pKa = 10.05IFASILYY54 pKa = 10.19LGGLGIGTGRR64 pKa = 11.84GTGGATGYY72 pKa = 11.23GPINPGGGRR81 pKa = 11.84ITGTGTVMRR90 pKa = 11.84PGVVIEE96 pKa = 4.19PVGPGDD102 pKa = 3.56IVSVGATDD110 pKa = 3.67SSIVPLLEE118 pKa = 4.09ATPDD122 pKa = 3.28IPIEE126 pKa = 4.48GGPEE130 pKa = 3.79VPPAGPDD137 pKa = 3.19VSTVDD142 pKa = 3.03VTANVDD148 pKa = 4.11PISDD152 pKa = 3.69INVTSSTTITNPDD165 pKa = 3.32SAVIDD170 pKa = 3.93VQPAPSGPRR179 pKa = 11.84RR180 pKa = 11.84VTVSRR185 pKa = 11.84SEE187 pKa = 4.07FQNASYY193 pKa = 11.39VSVTHH198 pKa = 6.8PSQGLGEE205 pKa = 4.33SGGAVISADD214 pKa = 3.1AGGSVVSSSHH224 pKa = 5.69EE225 pKa = 3.95LDD227 pKa = 3.23VGILIGGRR235 pKa = 11.84PPGNLSEE242 pKa = 4.23FQTIEE247 pKa = 3.89LDD249 pKa = 4.73EE250 pKa = 4.4IVQTQGVDD258 pKa = 3.23DD259 pKa = 4.27FDD261 pKa = 4.56IGEE264 pKa = 4.42GAGRR268 pKa = 11.84AGPDD272 pKa = 2.79TSTPDD277 pKa = 3.23NYY279 pKa = 10.81LGRR282 pKa = 11.84ALSRR286 pKa = 11.84FRR288 pKa = 11.84EE289 pKa = 4.31IYY291 pKa = 9.64EE292 pKa = 4.33RR293 pKa = 11.84PSDD296 pKa = 3.7LYY298 pKa = 9.96TRR300 pKa = 11.84RR301 pKa = 11.84VQQVRR306 pKa = 11.84VRR308 pKa = 11.84NRR310 pKa = 11.84DD311 pKa = 3.25LFLGDD316 pKa = 3.54PGRR319 pKa = 11.84LVTYY323 pKa = 10.03EE324 pKa = 3.81FDD326 pKa = 3.52NPAFEE331 pKa = 5.13GLGEE335 pKa = 4.03DD336 pKa = 4.41TIIFHH341 pKa = 6.45QAPEE345 pKa = 4.13VQAAPDD351 pKa = 4.09EE352 pKa = 4.47DD353 pKa = 4.3FMDD356 pKa = 4.9IIKK359 pKa = 10.7LGRR362 pKa = 11.84QRR364 pKa = 11.84LAQTPGGTVRR374 pKa = 11.84ISRR377 pKa = 11.84LGQRR381 pKa = 11.84GTIRR385 pKa = 11.84TRR387 pKa = 11.84SGLQIGGKK395 pKa = 7.39VHH397 pKa = 6.87FFSDD401 pKa = 4.05LSPIATEE408 pKa = 4.38NIEE411 pKa = 4.18LSPLGEE417 pKa = 3.97ISGEE421 pKa = 3.98AMVVDD426 pKa = 4.58ALSEE430 pKa = 3.98YY431 pKa = 11.25SLISEE436 pKa = 4.63PGIEE440 pKa = 4.44PVPFMDD446 pKa = 5.44EE447 pKa = 4.32DD448 pKa = 3.97LLDD451 pKa = 3.85IQSEE455 pKa = 4.34DD456 pKa = 3.57FSGSRR461 pKa = 11.84LHH463 pKa = 7.19LVRR466 pKa = 11.84TGNRR470 pKa = 11.84FGVIYY475 pKa = 10.06EE476 pKa = 3.9PSEE479 pKa = 3.78NLGARR484 pKa = 11.84TVFPDD489 pKa = 3.15INTGDD494 pKa = 4.9FIRR497 pKa = 11.84YY498 pKa = 5.82PQSNINPADD507 pKa = 3.29ISYY510 pKa = 11.08QDD512 pKa = 5.66LIPLQPGVDD521 pKa = 3.35INVGSIYY528 pKa = 10.24PSVDD532 pKa = 3.37YY533 pKa = 10.69YY534 pKa = 11.14LHH536 pKa = 6.65PSLRR540 pKa = 11.84RR541 pKa = 11.84RR542 pKa = 11.84KK543 pKa = 9.68RR544 pKa = 11.84KK545 pKa = 8.56RR546 pKa = 11.84TLHH549 pKa = 5.67
Molecular weight: 58.73 kDa
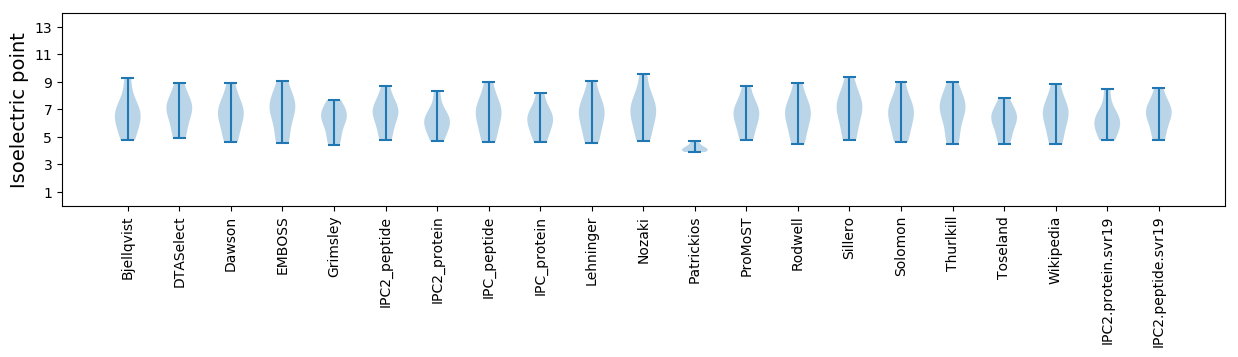
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|U6ELD1|U6ELD1_9PAPI Minor capsid protein L2 OS=Mesocricetus auratus papillomavirus 1 OX=1408129 GN=L2 PE=3 SV=1
MM1 pKa = 7.85AFPIEE6 pKa = 4.4LPALIQLLNTDD17 pKa = 3.1VTRR20 pKa = 11.84IVLYY24 pKa = 10.74CLFCDD29 pKa = 5.04RR30 pKa = 11.84PLSMQDD36 pKa = 2.65KK37 pKa = 10.15FDD39 pKa = 3.8YY40 pKa = 10.76FGAGLKK46 pKa = 9.02VVCRR50 pKa = 11.84KK51 pKa = 10.17GGMRR55 pKa = 11.84IACMSCRR62 pKa = 11.84RR63 pKa = 11.84ALAYY67 pKa = 10.4AEE69 pKa = 4.47DD70 pKa = 3.99LRR72 pKa = 11.84HH73 pKa = 5.57RR74 pKa = 11.84QCTAEE79 pKa = 4.12GDD81 pKa = 3.73LVEE84 pKa = 5.66RR85 pKa = 11.84ITGKK89 pKa = 10.35SLLHH93 pKa = 6.68IDD95 pKa = 3.88VRR97 pKa = 11.84CIACLCRR104 pKa = 11.84LGPSEE109 pKa = 4.8KK110 pKa = 10.52LLAKK114 pKa = 10.35ASHH117 pKa = 6.14KK118 pKa = 10.22PFYY121 pKa = 10.34LVRR124 pKa = 11.84SLWRR128 pKa = 11.84GPCRR132 pKa = 11.84KK133 pKa = 9.75CFLLL137 pKa = 5.23
MM1 pKa = 7.85AFPIEE6 pKa = 4.4LPALIQLLNTDD17 pKa = 3.1VTRR20 pKa = 11.84IVLYY24 pKa = 10.74CLFCDD29 pKa = 5.04RR30 pKa = 11.84PLSMQDD36 pKa = 2.65KK37 pKa = 10.15FDD39 pKa = 3.8YY40 pKa = 10.76FGAGLKK46 pKa = 9.02VVCRR50 pKa = 11.84KK51 pKa = 10.17GGMRR55 pKa = 11.84IACMSCRR62 pKa = 11.84RR63 pKa = 11.84ALAYY67 pKa = 10.4AEE69 pKa = 4.47DD70 pKa = 3.99LRR72 pKa = 11.84HH73 pKa = 5.57RR74 pKa = 11.84QCTAEE79 pKa = 4.12GDD81 pKa = 3.73LVEE84 pKa = 5.66RR85 pKa = 11.84ITGKK89 pKa = 10.35SLLHH93 pKa = 6.68IDD95 pKa = 3.88VRR97 pKa = 11.84CIACLCRR104 pKa = 11.84LGPSEE109 pKa = 4.8KK110 pKa = 10.52LLAKK114 pKa = 10.35ASHH117 pKa = 6.14KK118 pKa = 10.22PFYY121 pKa = 10.34LVRR124 pKa = 11.84SLWRR128 pKa = 11.84GPCRR132 pKa = 11.84KK133 pKa = 9.75CFLLL137 pKa = 5.23
Molecular weight: 15.58 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2514 |
137 |
608 |
359.1 |
40.02 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.444 ± 0.582 | 2.426 ± 0.707 |
6.762 ± 0.524 | 5.688 ± 0.643 |
4.097 ± 0.551 | 7.08 ± 1.129 |
1.671 ± 0.12 | 4.574 ± 0.797 |
4.733 ± 0.819 | 9.109 ± 0.623 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.71 ± 0.225 | 3.699 ± 0.62 |
7.518 ± 1.241 | 4.375 ± 0.566 |
6.484 ± 0.636 | 7.16 ± 0.659 |
5.927 ± 0.442 | 6.086 ± 0.749 |
1.034 ± 0.309 | 3.421 ± 0.404 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
