
Tortoise microvirus 91
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 7.06
Get precalculated fractions of proteins
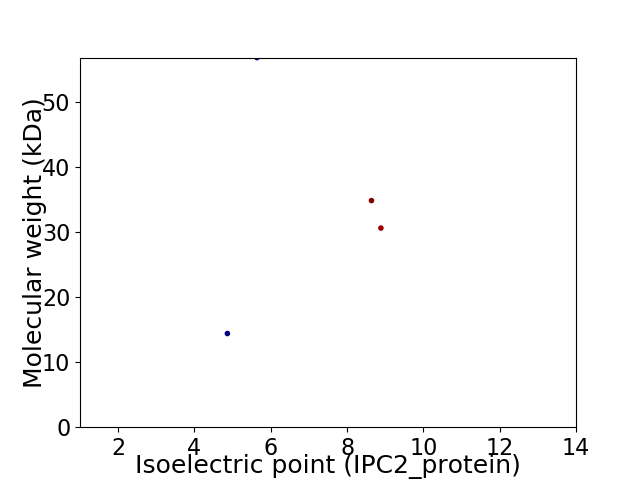
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4P8W725|A0A4P8W725_9VIRU Major capsid protein OS=Tortoise microvirus 91 OX=2583200 PE=3 SV=1
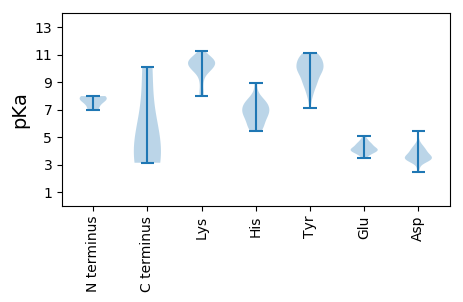
MM1 pKa = 6.99KK2 pKa = 9.45TIPFKK7 pKa = 10.9TPFNSIPDD15 pKa = 3.72EE16 pKa = 4.71GEE18 pKa = 4.05VNLDD22 pKa = 3.54PSEE25 pKa = 4.17TLPDD29 pKa = 3.4QSLSIMEE36 pKa = 4.09ILQRR40 pKa = 11.84HH41 pKa = 5.72AKK43 pKa = 9.77GLPLDD48 pKa = 3.54VKK50 pKa = 10.32MRR52 pKa = 11.84EE53 pKa = 4.29DD54 pKa = 3.58VGGGPDD60 pKa = 4.88DD61 pKa = 5.44FYY63 pKa = 10.97PDD65 pKa = 4.32PSTLDD70 pKa = 3.53LADD73 pKa = 4.31RR74 pKa = 11.84EE75 pKa = 4.48AFADD79 pKa = 3.7EE80 pKa = 4.51AKK82 pKa = 10.57RR83 pKa = 11.84EE84 pKa = 3.93LRR86 pKa = 11.84QIHH89 pKa = 6.97RR90 pKa = 11.84DD91 pKa = 3.21RR92 pKa = 11.84EE93 pKa = 3.84QRR95 pKa = 11.84KK96 pKa = 8.14QKK98 pKa = 10.51KK99 pKa = 9.92GSQKK103 pKa = 9.9PANPPKK109 pKa = 10.4KK110 pKa = 9.98DD111 pKa = 3.98DD112 pKa = 4.26PPKK115 pKa = 10.67KK116 pKa = 10.25DD117 pKa = 3.74DD118 pKa = 4.34TIQDD122 pKa = 3.66AEE124 pKa = 4.17IVEE127 pKa = 4.37
MM1 pKa = 6.99KK2 pKa = 9.45TIPFKK7 pKa = 10.9TPFNSIPDD15 pKa = 3.72EE16 pKa = 4.71GEE18 pKa = 4.05VNLDD22 pKa = 3.54PSEE25 pKa = 4.17TLPDD29 pKa = 3.4QSLSIMEE36 pKa = 4.09ILQRR40 pKa = 11.84HH41 pKa = 5.72AKK43 pKa = 9.77GLPLDD48 pKa = 3.54VKK50 pKa = 10.32MRR52 pKa = 11.84EE53 pKa = 4.29DD54 pKa = 3.58VGGGPDD60 pKa = 4.88DD61 pKa = 5.44FYY63 pKa = 10.97PDD65 pKa = 4.32PSTLDD70 pKa = 3.53LADD73 pKa = 4.31RR74 pKa = 11.84EE75 pKa = 4.48AFADD79 pKa = 3.7EE80 pKa = 4.51AKK82 pKa = 10.57RR83 pKa = 11.84EE84 pKa = 3.93LRR86 pKa = 11.84QIHH89 pKa = 6.97RR90 pKa = 11.84DD91 pKa = 3.21RR92 pKa = 11.84EE93 pKa = 3.84QRR95 pKa = 11.84KK96 pKa = 8.14QKK98 pKa = 10.51KK99 pKa = 9.92GSQKK103 pKa = 9.9PANPPKK109 pKa = 10.4KK110 pKa = 9.98DD111 pKa = 3.98DD112 pKa = 4.26PPKK115 pKa = 10.67KK116 pKa = 10.25DD117 pKa = 3.74DD118 pKa = 4.34TIQDD122 pKa = 3.66AEE124 pKa = 4.17IVEE127 pKa = 4.37
Molecular weight: 14.4 kDa
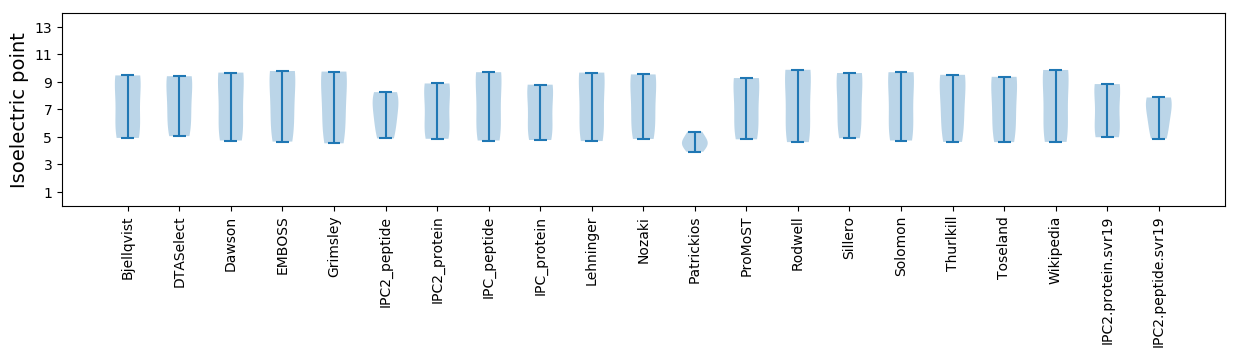
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4P8W6X6|A0A4P8W6X6_9VIRU Uncharacterized protein OS=Tortoise microvirus 91 OX=2583200 PE=4 SV=1
MM1 pKa = 7.7ACFTPMYY8 pKa = 10.15LEE10 pKa = 3.77QHH12 pKa = 5.54NVYY15 pKa = 9.56VPCGKK20 pKa = 10.5CPGCHH25 pKa = 6.01SARR28 pKa = 11.84ASAWSFRR35 pKa = 11.84MQQEE39 pKa = 4.31AKK41 pKa = 9.8ISTSAYY47 pKa = 9.72FVTLTYY53 pKa = 8.76DD54 pKa = 3.53TKK56 pKa = 10.84HH57 pKa = 6.79VPITPNGFMSLSKK70 pKa = 10.26RR71 pKa = 11.84DD72 pKa = 3.48CQLFFKK78 pKa = 10.52RR79 pKa = 11.84LRR81 pKa = 11.84KK82 pKa = 9.33NWPQTIRR89 pKa = 11.84YY90 pKa = 8.12YY91 pKa = 11.09LAGEE95 pKa = 4.2YY96 pKa = 10.46GDD98 pKa = 3.97SFNRR102 pKa = 11.84PHH104 pKa = 5.88YY105 pKa = 10.37HH106 pKa = 6.45VILFNSDD113 pKa = 3.05PQRR116 pKa = 11.84IVDD119 pKa = 3.19AWGNGDD125 pKa = 3.48VHH127 pKa = 7.89IADD130 pKa = 4.54VNPKK134 pKa = 8.08TCAYY138 pKa = 7.13TLKK141 pKa = 10.69YY142 pKa = 7.94ITKK145 pKa = 9.47GRR147 pKa = 11.84WKK149 pKa = 10.06PMHH152 pKa = 6.55KK153 pKa = 9.81RR154 pKa = 11.84DD155 pKa = 4.0DD156 pKa = 3.8RR157 pKa = 11.84APLFSLMSKK166 pKa = 10.74GLGKK170 pKa = 10.34NYY172 pKa = 8.85LTEE175 pKa = 5.05QMQDD179 pKa = 2.46WHH181 pKa = 6.71KK182 pKa = 11.18ANPAQRR188 pKa = 11.84VYY190 pKa = 9.8CTTSEE195 pKa = 4.35GVKK198 pKa = 8.84ITMPRR203 pKa = 11.84YY204 pKa = 9.17YY205 pKa = 10.04KK206 pKa = 10.67DD207 pKa = 3.21KK208 pKa = 10.36IYY210 pKa = 10.95SEE212 pKa = 4.55EE213 pKa = 3.83EE214 pKa = 3.46RR215 pKa = 11.84ALIAEE220 pKa = 4.77HH221 pKa = 6.68GLDD224 pKa = 3.47RR225 pKa = 11.84QTEE228 pKa = 4.18EE229 pKa = 4.06QRR231 pKa = 11.84KK232 pKa = 8.16QRR234 pKa = 11.84AANPNHH240 pKa = 6.61DD241 pKa = 4.5RR242 pKa = 11.84DD243 pKa = 3.74RR244 pKa = 11.84EE245 pKa = 4.21QYY247 pKa = 10.71VLNQFVKK254 pKa = 11.09AKK256 pKa = 8.02MTIKK260 pKa = 10.57KK261 pKa = 10.11
MM1 pKa = 7.7ACFTPMYY8 pKa = 10.15LEE10 pKa = 3.77QHH12 pKa = 5.54NVYY15 pKa = 9.56VPCGKK20 pKa = 10.5CPGCHH25 pKa = 6.01SARR28 pKa = 11.84ASAWSFRR35 pKa = 11.84MQQEE39 pKa = 4.31AKK41 pKa = 9.8ISTSAYY47 pKa = 9.72FVTLTYY53 pKa = 8.76DD54 pKa = 3.53TKK56 pKa = 10.84HH57 pKa = 6.79VPITPNGFMSLSKK70 pKa = 10.26RR71 pKa = 11.84DD72 pKa = 3.48CQLFFKK78 pKa = 10.52RR79 pKa = 11.84LRR81 pKa = 11.84KK82 pKa = 9.33NWPQTIRR89 pKa = 11.84YY90 pKa = 8.12YY91 pKa = 11.09LAGEE95 pKa = 4.2YY96 pKa = 10.46GDD98 pKa = 3.97SFNRR102 pKa = 11.84PHH104 pKa = 5.88YY105 pKa = 10.37HH106 pKa = 6.45VILFNSDD113 pKa = 3.05PQRR116 pKa = 11.84IVDD119 pKa = 3.19AWGNGDD125 pKa = 3.48VHH127 pKa = 7.89IADD130 pKa = 4.54VNPKK134 pKa = 8.08TCAYY138 pKa = 7.13TLKK141 pKa = 10.69YY142 pKa = 7.94ITKK145 pKa = 9.47GRR147 pKa = 11.84WKK149 pKa = 10.06PMHH152 pKa = 6.55KK153 pKa = 9.81RR154 pKa = 11.84DD155 pKa = 4.0DD156 pKa = 3.8RR157 pKa = 11.84APLFSLMSKK166 pKa = 10.74GLGKK170 pKa = 10.34NYY172 pKa = 8.85LTEE175 pKa = 5.05QMQDD179 pKa = 2.46WHH181 pKa = 6.71KK182 pKa = 11.18ANPAQRR188 pKa = 11.84VYY190 pKa = 9.8CTTSEE195 pKa = 4.35GVKK198 pKa = 8.84ITMPRR203 pKa = 11.84YY204 pKa = 9.17YY205 pKa = 10.04KK206 pKa = 10.67DD207 pKa = 3.21KK208 pKa = 10.36IYY210 pKa = 10.95SEE212 pKa = 4.55EE213 pKa = 3.83EE214 pKa = 3.46RR215 pKa = 11.84ALIAEE220 pKa = 4.77HH221 pKa = 6.68GLDD224 pKa = 3.47RR225 pKa = 11.84QTEE228 pKa = 4.18EE229 pKa = 4.06QRR231 pKa = 11.84KK232 pKa = 8.16QRR234 pKa = 11.84AANPNHH240 pKa = 6.61DD241 pKa = 4.5RR242 pKa = 11.84DD243 pKa = 3.74RR244 pKa = 11.84EE245 pKa = 4.21QYY247 pKa = 10.71VLNQFVKK254 pKa = 11.09AKK256 pKa = 8.02MTIKK260 pKa = 10.57KK261 pKa = 10.11
Molecular weight: 30.62 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1203 |
127 |
503 |
300.8 |
34.18 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.063 ± 0.978 | 0.831 ± 0.52 |
5.736 ± 1.259 | 5.902 ± 0.558 |
3.99 ± 0.632 | 5.819 ± 0.586 |
2.078 ± 0.594 | 5.486 ± 0.413 |
5.736 ± 1.252 | 7.232 ± 0.925 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.741 ± 0.278 | 5.902 ± 1.377 |
6.401 ± 1.365 | 5.736 ± 1.526 |
6.318 ± 0.705 | 5.569 ± 0.264 |
5.985 ± 0.534 | 4.239 ± 0.334 |
1.164 ± 0.47 | 4.073 ± 0.975 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
