
Picobirnavirus Equ3
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Duplopiviricetes; Durnavirales; Picobirnaviridae; Picobirnavirus; unclassified Picobirnavirus
Average proteome isoelectric point is 5.45
Get precalculated fractions of proteins
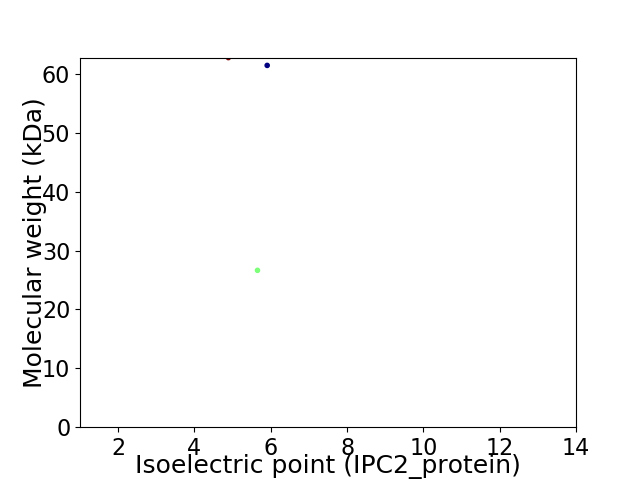
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0H4ATH8|A0A0H4ATH8_9VIRU ORF1 OS=Picobirnavirus Equ3 OX=1673646 PE=4 SV=1
MM1 pKa = 7.49KK2 pKa = 10.74GEE4 pKa = 3.92LTMANNNNHH13 pKa = 5.75SKK15 pKa = 10.17NRR17 pKa = 11.84RR18 pKa = 11.84GNEE21 pKa = 3.55EE22 pKa = 3.91NEE24 pKa = 4.37EE25 pKa = 4.01KK26 pKa = 10.6RR27 pKa = 11.84SSKK30 pKa = 9.66KK31 pKa = 10.29DD32 pKa = 3.06KK33 pKa = 10.39RR34 pKa = 11.84ISYY37 pKa = 9.57NAYY40 pKa = 9.13KK41 pKa = 10.48SKK43 pKa = 10.85PNDD46 pKa = 3.54PSWYY50 pKa = 6.73EE51 pKa = 3.87TTPEE55 pKa = 3.72LTKK58 pKa = 10.9AVTNFSFDD66 pKa = 3.15TATGYY71 pKa = 11.35AFGMGDD77 pKa = 4.73LDD79 pKa = 5.14DD80 pKa = 5.1PMHH83 pKa = 6.05GTSNPGIMTIAVSPTVGWATNPNSPVNNAAMHH115 pKa = 5.86VYY117 pKa = 10.63SFIRR121 pKa = 11.84HH122 pKa = 5.29EE123 pKa = 4.0NSGNKK128 pKa = 10.03NYY130 pKa = 10.69DD131 pKa = 3.32PNDD134 pKa = 3.25YY135 pKa = 10.67FLYY138 pKa = 10.51LLSYY142 pKa = 10.71DD143 pKa = 4.76GISMWLEE150 pKa = 3.44FLKK153 pKa = 10.51RR154 pKa = 11.84VYY156 pKa = 10.59GVANTYY162 pKa = 10.06HH163 pKa = 6.63PHH165 pKa = 6.01NEE167 pKa = 3.94YY168 pKa = 10.37FPRR171 pKa = 11.84AVLTAMGINAKK182 pKa = 10.3DD183 pKa = 3.59IIKK186 pKa = 10.31HH187 pKa = 5.16KK188 pKa = 10.34PRR190 pKa = 11.84LLTLINTTALDD201 pKa = 3.91LSLKK205 pKa = 10.05KK206 pKa = 10.82VPAYY210 pKa = 10.43LPYY213 pKa = 9.95GAKK216 pKa = 10.02HH217 pKa = 5.42RR218 pKa = 11.84WLMEE222 pKa = 3.56GVYY225 pKa = 10.38QDD227 pKa = 4.49ANLAKK232 pKa = 8.78AQVYY236 pKa = 10.31AFVPEE241 pKa = 4.42YY242 pKa = 10.42IFEE245 pKa = 4.1YY246 pKa = 10.85DD247 pKa = 3.27IDD249 pKa = 4.17EE250 pKa = 5.13DD251 pKa = 4.21GAGCVVPKK259 pKa = 10.25PFYY262 pKa = 10.79SQALALGGYY271 pKa = 9.65GIEE274 pKa = 4.17EE275 pKa = 4.91LEE277 pKa = 4.41TFTQNLMDD285 pKa = 3.61SFFRR289 pKa = 11.84EE290 pKa = 3.48EE291 pKa = 4.25DD292 pKa = 3.5LGVMSGDD299 pKa = 3.16TMKK302 pKa = 10.76AYY304 pKa = 10.29GSNCYY309 pKa = 10.54APMGIEE315 pKa = 3.46STYY318 pKa = 10.87RR319 pKa = 11.84VLPSYY324 pKa = 10.6LPEE327 pKa = 4.34VLDD330 pKa = 4.16QIQNLTLMGGYY341 pKa = 10.16DD342 pKa = 3.6GATCRR347 pKa = 11.84VVQSEE352 pKa = 4.24DD353 pKa = 3.0KK354 pKa = 10.59DD355 pKa = 3.99YY356 pKa = 11.48LVSKK360 pKa = 9.51PEE362 pKa = 3.43WRR364 pKa = 11.84LPEE367 pKa = 4.17EE368 pKa = 4.0YY369 pKa = 9.83RR370 pKa = 11.84AGGRR374 pKa = 11.84TPFMRR379 pKa = 11.84EE380 pKa = 3.17QFLNFEE386 pKa = 4.38MAAVNHH392 pKa = 6.26GNVAEE397 pKa = 4.22ATRR400 pKa = 11.84MTNIASVYY408 pKa = 10.53NEE410 pKa = 4.01VEE412 pKa = 3.92NTIAATTIGSEE423 pKa = 4.09VANYY427 pKa = 9.91AVIWTFGYY435 pKa = 10.67NEE437 pKa = 4.91NIGQFDD443 pKa = 3.92LHH445 pKa = 5.84RR446 pKa = 11.84TPAIFTSFLAPIKK459 pKa = 10.36ADD461 pKa = 3.46SGHH464 pKa = 7.07SLMGMIDD471 pKa = 3.58ADD473 pKa = 3.89VVVNALQPFMYY484 pKa = 9.83DD485 pKa = 4.42LSVLCQFDD493 pKa = 3.81RR494 pKa = 11.84HH495 pKa = 4.95PQIYY499 pKa = 7.74ATAYY503 pKa = 9.74VSYY506 pKa = 9.77PNPDD510 pKa = 3.62DD511 pKa = 4.93PEE513 pKa = 4.67TDD515 pKa = 3.29VTKK518 pKa = 10.15WVYY521 pKa = 10.92ARR523 pKa = 11.84NFLGDD528 pKa = 3.67LANYY532 pKa = 9.04YY533 pKa = 10.82VIDD536 pKa = 3.76RR537 pKa = 11.84TEE539 pKa = 4.51LEE541 pKa = 4.24LLSTNALKK549 pKa = 11.03AEE551 pKa = 4.46FGLMTNN557 pKa = 4.23
MM1 pKa = 7.49KK2 pKa = 10.74GEE4 pKa = 3.92LTMANNNNHH13 pKa = 5.75SKK15 pKa = 10.17NRR17 pKa = 11.84RR18 pKa = 11.84GNEE21 pKa = 3.55EE22 pKa = 3.91NEE24 pKa = 4.37EE25 pKa = 4.01KK26 pKa = 10.6RR27 pKa = 11.84SSKK30 pKa = 9.66KK31 pKa = 10.29DD32 pKa = 3.06KK33 pKa = 10.39RR34 pKa = 11.84ISYY37 pKa = 9.57NAYY40 pKa = 9.13KK41 pKa = 10.48SKK43 pKa = 10.85PNDD46 pKa = 3.54PSWYY50 pKa = 6.73EE51 pKa = 3.87TTPEE55 pKa = 3.72LTKK58 pKa = 10.9AVTNFSFDD66 pKa = 3.15TATGYY71 pKa = 11.35AFGMGDD77 pKa = 4.73LDD79 pKa = 5.14DD80 pKa = 5.1PMHH83 pKa = 6.05GTSNPGIMTIAVSPTVGWATNPNSPVNNAAMHH115 pKa = 5.86VYY117 pKa = 10.63SFIRR121 pKa = 11.84HH122 pKa = 5.29EE123 pKa = 4.0NSGNKK128 pKa = 10.03NYY130 pKa = 10.69DD131 pKa = 3.32PNDD134 pKa = 3.25YY135 pKa = 10.67FLYY138 pKa = 10.51LLSYY142 pKa = 10.71DD143 pKa = 4.76GISMWLEE150 pKa = 3.44FLKK153 pKa = 10.51RR154 pKa = 11.84VYY156 pKa = 10.59GVANTYY162 pKa = 10.06HH163 pKa = 6.63PHH165 pKa = 6.01NEE167 pKa = 3.94YY168 pKa = 10.37FPRR171 pKa = 11.84AVLTAMGINAKK182 pKa = 10.3DD183 pKa = 3.59IIKK186 pKa = 10.31HH187 pKa = 5.16KK188 pKa = 10.34PRR190 pKa = 11.84LLTLINTTALDD201 pKa = 3.91LSLKK205 pKa = 10.05KK206 pKa = 10.82VPAYY210 pKa = 10.43LPYY213 pKa = 9.95GAKK216 pKa = 10.02HH217 pKa = 5.42RR218 pKa = 11.84WLMEE222 pKa = 3.56GVYY225 pKa = 10.38QDD227 pKa = 4.49ANLAKK232 pKa = 8.78AQVYY236 pKa = 10.31AFVPEE241 pKa = 4.42YY242 pKa = 10.42IFEE245 pKa = 4.1YY246 pKa = 10.85DD247 pKa = 3.27IDD249 pKa = 4.17EE250 pKa = 5.13DD251 pKa = 4.21GAGCVVPKK259 pKa = 10.25PFYY262 pKa = 10.79SQALALGGYY271 pKa = 9.65GIEE274 pKa = 4.17EE275 pKa = 4.91LEE277 pKa = 4.41TFTQNLMDD285 pKa = 3.61SFFRR289 pKa = 11.84EE290 pKa = 3.48EE291 pKa = 4.25DD292 pKa = 3.5LGVMSGDD299 pKa = 3.16TMKK302 pKa = 10.76AYY304 pKa = 10.29GSNCYY309 pKa = 10.54APMGIEE315 pKa = 3.46STYY318 pKa = 10.87RR319 pKa = 11.84VLPSYY324 pKa = 10.6LPEE327 pKa = 4.34VLDD330 pKa = 4.16QIQNLTLMGGYY341 pKa = 10.16DD342 pKa = 3.6GATCRR347 pKa = 11.84VVQSEE352 pKa = 4.24DD353 pKa = 3.0KK354 pKa = 10.59DD355 pKa = 3.99YY356 pKa = 11.48LVSKK360 pKa = 9.51PEE362 pKa = 3.43WRR364 pKa = 11.84LPEE367 pKa = 4.17EE368 pKa = 4.0YY369 pKa = 9.83RR370 pKa = 11.84AGGRR374 pKa = 11.84TPFMRR379 pKa = 11.84EE380 pKa = 3.17QFLNFEE386 pKa = 4.38MAAVNHH392 pKa = 6.26GNVAEE397 pKa = 4.22ATRR400 pKa = 11.84MTNIASVYY408 pKa = 10.53NEE410 pKa = 4.01VEE412 pKa = 3.92NTIAATTIGSEE423 pKa = 4.09VANYY427 pKa = 9.91AVIWTFGYY435 pKa = 10.67NEE437 pKa = 4.91NIGQFDD443 pKa = 3.92LHH445 pKa = 5.84RR446 pKa = 11.84TPAIFTSFLAPIKK459 pKa = 10.36ADD461 pKa = 3.46SGHH464 pKa = 7.07SLMGMIDD471 pKa = 3.58ADD473 pKa = 3.89VVVNALQPFMYY484 pKa = 9.83DD485 pKa = 4.42LSVLCQFDD493 pKa = 3.81RR494 pKa = 11.84HH495 pKa = 4.95PQIYY499 pKa = 7.74ATAYY503 pKa = 9.74VSYY506 pKa = 9.77PNPDD510 pKa = 3.62DD511 pKa = 4.93PEE513 pKa = 4.67TDD515 pKa = 3.29VTKK518 pKa = 10.15WVYY521 pKa = 10.92ARR523 pKa = 11.84NFLGDD528 pKa = 3.67LANYY532 pKa = 9.04YY533 pKa = 10.82VIDD536 pKa = 3.76RR537 pKa = 11.84TEE539 pKa = 4.51LEE541 pKa = 4.24LLSTNALKK549 pKa = 11.03AEE551 pKa = 4.46FGLMTNN557 pKa = 4.23
Molecular weight: 62.78 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0H4AQ74|A0A0H4AQ74_9VIRU Capsid protein OS=Picobirnavirus Equ3 OX=1673646 PE=4 SV=1
MM1 pKa = 7.62KK2 pKa = 10.14IEE4 pKa = 4.27SLPSEE9 pKa = 4.59VYY11 pKa = 10.59DD12 pKa = 4.03VVLNNDD18 pKa = 3.75GLKK21 pKa = 9.66MYY23 pKa = 10.67LNNLEE28 pKa = 4.49RR29 pKa = 11.84GRR31 pKa = 11.84SATPRR36 pKa = 11.84SWLYY40 pKa = 10.04EE41 pKa = 4.04GKK43 pKa = 10.17SDD45 pKa = 5.04TEE47 pKa = 4.49VLQAWLTILEE57 pKa = 4.51TVKK60 pKa = 10.74SSEE63 pKa = 4.11FGEE66 pKa = 5.05LIYY69 pKa = 10.9QFDD72 pKa = 3.78TSQLKK77 pKa = 9.76KK78 pKa = 9.56WGAQGEE84 pKa = 4.47VKK86 pKa = 10.18PIKK89 pKa = 10.07EE90 pKa = 3.87LMEE93 pKa = 4.07IVTEE97 pKa = 4.31GFTNAGTPKK106 pKa = 9.5PAIWNTPMWQQAKK119 pKa = 9.24TNAIKK124 pKa = 10.83YY125 pKa = 9.04LIIDD129 pKa = 3.5TGLYY133 pKa = 9.75KK134 pKa = 10.54RR135 pKa = 11.84LRR137 pKa = 11.84PRR139 pKa = 11.84ALSHH143 pKa = 6.55VVDD146 pKa = 4.82DD147 pKa = 3.95MRR149 pKa = 11.84DD150 pKa = 3.47RR151 pKa = 11.84DD152 pKa = 4.13TLEE155 pKa = 4.43SNSGFPDD162 pKa = 4.05FGRR165 pKa = 11.84RR166 pKa = 11.84KK167 pKa = 9.98KK168 pKa = 10.69PEE170 pKa = 3.57ILTAAFEE177 pKa = 4.74AIKK180 pKa = 10.74SGEE183 pKa = 3.93FWEE186 pKa = 5.61FPAIVLFRR194 pKa = 11.84NYY196 pKa = 8.69NQKK199 pKa = 8.3TRR201 pKa = 11.84IVWMYY206 pKa = 10.73PMATNIYY213 pKa = 8.93EE214 pKa = 4.42GSFTQPLKK222 pKa = 9.86EE223 pKa = 4.19ALMKK227 pKa = 10.56SDD229 pKa = 5.31LDD231 pKa = 5.55FIAPWRR237 pKa = 11.84GYY239 pKa = 10.15DD240 pKa = 3.28HH241 pKa = 7.0VLARR245 pKa = 11.84LSKK248 pKa = 10.72LYY250 pKa = 9.29GTGEE254 pKa = 4.12FLSASDD260 pKa = 4.72FSHH263 pKa = 6.94TDD265 pKa = 2.65AHH267 pKa = 5.21FTKK270 pKa = 10.04WAILEE275 pKa = 4.23VYY277 pKa = 10.29DD278 pKa = 3.89VIKK281 pKa = 10.93YY282 pKa = 9.78AFQEE286 pKa = 4.38QYY288 pKa = 9.78WDD290 pKa = 3.46EE291 pKa = 4.48LKK293 pKa = 10.81RR294 pKa = 11.84SMLHH298 pKa = 5.01VVEE301 pKa = 4.72IPLIIGEE308 pKa = 4.2NSWIIGDD315 pKa = 4.19HH316 pKa = 5.76GVSSGSNWTNDD327 pKa = 2.56VEE329 pKa = 4.67TYY331 pKa = 9.62MDD333 pKa = 5.01FIIAQYY339 pKa = 9.06LTLLKK344 pKa = 10.35LVKK347 pKa = 9.24EE348 pKa = 4.19PCMAIGDD355 pKa = 4.77DD356 pKa = 3.27ISHH359 pKa = 6.65RR360 pKa = 11.84RR361 pKa = 11.84DD362 pKa = 3.1SYY364 pKa = 11.79LPDD367 pKa = 4.34LDD369 pKa = 3.97EE370 pKa = 6.23QIAQVCQSMGFDD382 pKa = 3.43VNAEE386 pKa = 4.15KK387 pKa = 9.95VTNQQDD393 pKa = 3.24WVKK396 pKa = 10.1FLQRR400 pKa = 11.84LTIRR404 pKa = 11.84GFNSEE409 pKa = 3.49RR410 pKa = 11.84TYY412 pKa = 11.0SVEE415 pKa = 3.71GVEE418 pKa = 4.25YY419 pKa = 9.74PLLRR423 pKa = 11.84GIYY426 pKa = 8.39STIRR430 pKa = 11.84ALNSSLNPEE439 pKa = 4.47KK440 pKa = 10.57FHH442 pKa = 7.59SPKK445 pKa = 10.38LWSKK449 pKa = 11.74DD450 pKa = 3.28MFAVRR455 pKa = 11.84QFMILEE461 pKa = 4.07NCIDD465 pKa = 3.82HH466 pKa = 6.94PLFVQFVKK474 pKa = 9.7FVCAGNPYY482 pKa = 10.04LVKK485 pKa = 10.2FAKK488 pKa = 10.29LKK490 pKa = 10.9NEE492 pKa = 4.15QIDD495 pKa = 4.1KK496 pKa = 10.56KK497 pKa = 9.1WAQSRR502 pKa = 11.84ILPGLNPTYY511 pKa = 10.41NQEE514 pKa = 4.26KK515 pKa = 9.62RR516 pKa = 11.84DD517 pKa = 4.09KK518 pKa = 10.65PLSTYY523 pKa = 10.79AAIAIARR530 pKa = 11.84SLL532 pKa = 3.55
MM1 pKa = 7.62KK2 pKa = 10.14IEE4 pKa = 4.27SLPSEE9 pKa = 4.59VYY11 pKa = 10.59DD12 pKa = 4.03VVLNNDD18 pKa = 3.75GLKK21 pKa = 9.66MYY23 pKa = 10.67LNNLEE28 pKa = 4.49RR29 pKa = 11.84GRR31 pKa = 11.84SATPRR36 pKa = 11.84SWLYY40 pKa = 10.04EE41 pKa = 4.04GKK43 pKa = 10.17SDD45 pKa = 5.04TEE47 pKa = 4.49VLQAWLTILEE57 pKa = 4.51TVKK60 pKa = 10.74SSEE63 pKa = 4.11FGEE66 pKa = 5.05LIYY69 pKa = 10.9QFDD72 pKa = 3.78TSQLKK77 pKa = 9.76KK78 pKa = 9.56WGAQGEE84 pKa = 4.47VKK86 pKa = 10.18PIKK89 pKa = 10.07EE90 pKa = 3.87LMEE93 pKa = 4.07IVTEE97 pKa = 4.31GFTNAGTPKK106 pKa = 9.5PAIWNTPMWQQAKK119 pKa = 9.24TNAIKK124 pKa = 10.83YY125 pKa = 9.04LIIDD129 pKa = 3.5TGLYY133 pKa = 9.75KK134 pKa = 10.54RR135 pKa = 11.84LRR137 pKa = 11.84PRR139 pKa = 11.84ALSHH143 pKa = 6.55VVDD146 pKa = 4.82DD147 pKa = 3.95MRR149 pKa = 11.84DD150 pKa = 3.47RR151 pKa = 11.84DD152 pKa = 4.13TLEE155 pKa = 4.43SNSGFPDD162 pKa = 4.05FGRR165 pKa = 11.84RR166 pKa = 11.84KK167 pKa = 9.98KK168 pKa = 10.69PEE170 pKa = 3.57ILTAAFEE177 pKa = 4.74AIKK180 pKa = 10.74SGEE183 pKa = 3.93FWEE186 pKa = 5.61FPAIVLFRR194 pKa = 11.84NYY196 pKa = 8.69NQKK199 pKa = 8.3TRR201 pKa = 11.84IVWMYY206 pKa = 10.73PMATNIYY213 pKa = 8.93EE214 pKa = 4.42GSFTQPLKK222 pKa = 9.86EE223 pKa = 4.19ALMKK227 pKa = 10.56SDD229 pKa = 5.31LDD231 pKa = 5.55FIAPWRR237 pKa = 11.84GYY239 pKa = 10.15DD240 pKa = 3.28HH241 pKa = 7.0VLARR245 pKa = 11.84LSKK248 pKa = 10.72LYY250 pKa = 9.29GTGEE254 pKa = 4.12FLSASDD260 pKa = 4.72FSHH263 pKa = 6.94TDD265 pKa = 2.65AHH267 pKa = 5.21FTKK270 pKa = 10.04WAILEE275 pKa = 4.23VYY277 pKa = 10.29DD278 pKa = 3.89VIKK281 pKa = 10.93YY282 pKa = 9.78AFQEE286 pKa = 4.38QYY288 pKa = 9.78WDD290 pKa = 3.46EE291 pKa = 4.48LKK293 pKa = 10.81RR294 pKa = 11.84SMLHH298 pKa = 5.01VVEE301 pKa = 4.72IPLIIGEE308 pKa = 4.2NSWIIGDD315 pKa = 4.19HH316 pKa = 5.76GVSSGSNWTNDD327 pKa = 2.56VEE329 pKa = 4.67TYY331 pKa = 9.62MDD333 pKa = 5.01FIIAQYY339 pKa = 9.06LTLLKK344 pKa = 10.35LVKK347 pKa = 9.24EE348 pKa = 4.19PCMAIGDD355 pKa = 4.77DD356 pKa = 3.27ISHH359 pKa = 6.65RR360 pKa = 11.84RR361 pKa = 11.84DD362 pKa = 3.1SYY364 pKa = 11.79LPDD367 pKa = 4.34LDD369 pKa = 3.97EE370 pKa = 6.23QIAQVCQSMGFDD382 pKa = 3.43VNAEE386 pKa = 4.15KK387 pKa = 9.95VTNQQDD393 pKa = 3.24WVKK396 pKa = 10.1FLQRR400 pKa = 11.84LTIRR404 pKa = 11.84GFNSEE409 pKa = 3.49RR410 pKa = 11.84TYY412 pKa = 11.0SVEE415 pKa = 3.71GVEE418 pKa = 4.25YY419 pKa = 9.74PLLRR423 pKa = 11.84GIYY426 pKa = 8.39STIRR430 pKa = 11.84ALNSSLNPEE439 pKa = 4.47KK440 pKa = 10.57FHH442 pKa = 7.59SPKK445 pKa = 10.38LWSKK449 pKa = 11.74DD450 pKa = 3.28MFAVRR455 pKa = 11.84QFMILEE461 pKa = 4.07NCIDD465 pKa = 3.82HH466 pKa = 6.94PLFVQFVKK474 pKa = 9.7FVCAGNPYY482 pKa = 10.04LVKK485 pKa = 10.2FAKK488 pKa = 10.29LKK490 pKa = 10.9NEE492 pKa = 4.15QIDD495 pKa = 4.1KK496 pKa = 10.56KK497 pKa = 9.1WAQSRR502 pKa = 11.84ILPGLNPTYY511 pKa = 10.41NQEE514 pKa = 4.26KK515 pKa = 9.62RR516 pKa = 11.84DD517 pKa = 4.09KK518 pKa = 10.65PLSTYY523 pKa = 10.79AAIAIARR530 pKa = 11.84SLL532 pKa = 3.55
Molecular weight: 61.45 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1340 |
251 |
557 |
446.7 |
50.29 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.955 ± 2.272 | 0.597 ± 0.184 |
5.373 ± 0.672 | 6.716 ± 0.162 |
3.881 ± 0.971 | 6.642 ± 1.23 |
2.015 ± 0.181 | 5.224 ± 0.825 |
5.299 ± 0.909 | 7.985 ± 1.121 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.985 ± 0.44 | 6.716 ± 1.269 |
4.925 ± 0.256 | 3.507 ± 0.755 |
4.478 ± 0.382 | 6.194 ± 0.351 |
5.97 ± 0.401 | 5.448 ± 0.583 |
2.015 ± 0.455 | 5.075 ± 0.87 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
