
Leshenault partiti-like virus
Taxonomy: Viruses; unclassified viruses
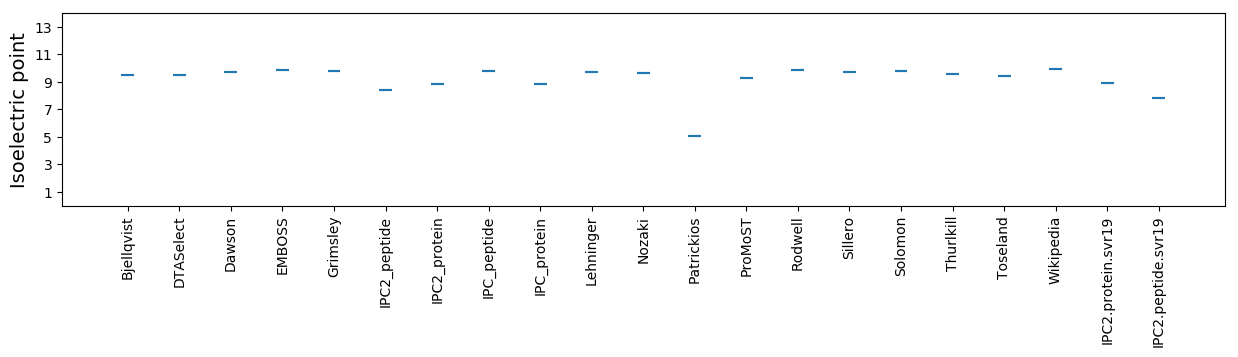
Average proteome isoelectric point is 8.92
Get precalculated fractions of proteins
Virtual 2D-PAGE plot for 1 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
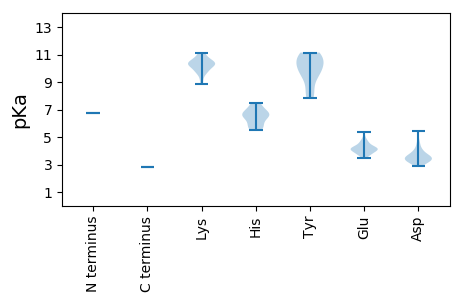
Protein with the lowest isoelectric point:
>tr|A0A1Z2RT46|A0A1Z2RT46_9VIRU RdRp OS=Leshenault partiti-like virus OX=2010278 PE=4 SV=1
MM1 pKa = 6.75QASRR5 pKa = 11.84MLEE8 pKa = 3.8SHH10 pKa = 6.39RR11 pKa = 11.84VWGEE15 pKa = 3.59YY16 pKa = 7.87RR17 pKa = 11.84THH19 pKa = 7.16RR20 pKa = 11.84LLEE23 pKa = 4.22AHH25 pKa = 7.37DD26 pKa = 5.42GRR28 pKa = 11.84ATWLRR33 pKa = 11.84AWTKK37 pKa = 10.23SWAPRR42 pKa = 11.84QGVVEE47 pKa = 4.18STPEE51 pKa = 3.78AEE53 pKa = 4.01KK54 pKa = 10.7HH55 pKa = 5.5VGVMTYY61 pKa = 10.32VKK63 pKa = 9.87GWRR66 pKa = 11.84RR67 pKa = 11.84RR68 pKa = 11.84LRR70 pKa = 11.84RR71 pKa = 11.84QLRR74 pKa = 11.84SARR77 pKa = 11.84TQFEE81 pKa = 4.24RR82 pKa = 11.84EE83 pKa = 4.19VVHH86 pKa = 6.55GSWAALEE93 pKa = 4.18GLYY96 pKa = 10.49SEE98 pKa = 5.4ALEE101 pKa = 4.81HH102 pKa = 6.64SPTMDD107 pKa = 3.24LMYY110 pKa = 11.0GHH112 pKa = 6.75VLSLSVDD119 pKa = 3.49RR120 pKa = 11.84DD121 pKa = 3.41HH122 pKa = 7.5GFTRR126 pKa = 11.84SFPSWYY132 pKa = 9.64VSPAGFEE139 pKa = 3.78NVPIVKK145 pKa = 8.82RR146 pKa = 11.84TTAKK150 pKa = 10.9DD151 pKa = 3.1PFTRR155 pKa = 11.84MVEE158 pKa = 4.04TGPTGIVARR167 pKa = 11.84TMMKK171 pKa = 10.29VGGRR175 pKa = 11.84PQKK178 pKa = 10.03FTDD181 pKa = 3.43AAATSLAYY189 pKa = 9.41KK190 pKa = 9.21WLHH193 pKa = 5.59ATGLWSSVTDD203 pKa = 3.67ADD205 pKa = 4.04VVINVSASFDD215 pKa = 3.4TSRR218 pKa = 11.84VHH220 pKa = 5.78MEE222 pKa = 4.03KK223 pKa = 9.68FTAPRR228 pKa = 11.84PPTEE232 pKa = 4.29GTLDD236 pKa = 3.55MCWEE240 pKa = 4.21VVARR244 pKa = 11.84NFKK247 pKa = 9.76EE248 pKa = 4.0LGMTGLPWITVADD261 pKa = 4.65VIAPHH266 pKa = 6.76INGDD270 pKa = 3.7ACPGLLTSRR279 pKa = 11.84IYY281 pKa = 9.73RR282 pKa = 11.84TRR284 pKa = 11.84AQASSCTTSDD294 pKa = 3.21VVALFEE300 pKa = 4.37ALRR303 pKa = 11.84TQEE306 pKa = 4.73LLCALPWAAGGRR318 pKa = 11.84EE319 pKa = 3.79KK320 pKa = 10.69RR321 pKa = 11.84QEE323 pKa = 4.12RR324 pKa = 11.84LVGDD328 pKa = 3.72QVSSRR333 pKa = 11.84LVQMPEE339 pKa = 3.76DD340 pKa = 3.38VLARR344 pKa = 11.84LEE346 pKa = 4.27SAVAGPLLQRR356 pKa = 11.84LKK358 pKa = 9.8WVKK361 pKa = 10.39RR362 pKa = 11.84DD363 pKa = 3.16IALGMSFFRR372 pKa = 11.84GRR374 pKa = 11.84AQRR377 pKa = 11.84FFHH380 pKa = 7.19RR381 pKa = 11.84FDD383 pKa = 3.17TCTHH387 pKa = 5.6VKK389 pKa = 10.49SFDD392 pKa = 2.93WSAFDD397 pKa = 3.61SSVGEE402 pKa = 4.12TLIVASFSLLRR413 pKa = 11.84ACYY416 pKa = 9.9PEE418 pKa = 4.72GQHH421 pKa = 6.6ADD423 pKa = 3.31NVFKK427 pKa = 11.13YY428 pKa = 9.68ILSAFVHH435 pKa = 5.85KK436 pKa = 10.29HH437 pKa = 5.92LIVPQGDD444 pKa = 4.06VIGMHH449 pKa = 6.74RR450 pKa = 11.84GIPSGSGLTSLVGSVANWLILRR472 pKa = 11.84TTLTDD477 pKa = 3.28ICPPLLAQRR486 pKa = 11.84AEE488 pKa = 4.22VAVLGDD494 pKa = 3.42DD495 pKa = 3.58TLIGFRR501 pKa = 11.84GYY503 pKa = 11.14DD504 pKa = 3.99GIPTNAEE511 pKa = 3.44FTARR515 pKa = 11.84AFALWGVAAGPSPSMVYY532 pKa = 10.35PEE534 pKa = 4.51GPPRR538 pKa = 11.84SVYY541 pKa = 10.5LEE543 pKa = 3.97RR544 pKa = 11.84CQTFLSVTSWCGLPSWRR561 pKa = 11.84NRR563 pKa = 11.84EE564 pKa = 3.66WWVMDD569 pKa = 3.04MGYY572 pKa = 11.08AKK574 pKa = 10.28GQPDD578 pKa = 3.47VANAQVAALEE588 pKa = 4.35DD589 pKa = 3.65AGFSGFAPQLRR600 pKa = 11.84GTRR603 pKa = 11.84RR604 pKa = 11.84RR605 pKa = 11.84FQKK608 pKa = 9.66WLQKK612 pKa = 10.53QPGAHH617 pKa = 6.2MLHH620 pKa = 6.83PEE622 pKa = 4.12YY623 pKa = 11.08VEE625 pKa = 4.02STEE628 pKa = 4.54TITQGWMARR637 pKa = 11.84AHH639 pKa = 6.41SSTGTAKK646 pKa = 10.35EE647 pKa = 3.69LAAFDD652 pKa = 4.07WDD654 pKa = 4.49SKK656 pKa = 10.39GQHH659 pKa = 6.38PGNWPTRR666 pKa = 11.84WKK668 pKa = 10.34VAAEE672 pKa = 3.99AAPPPHH678 pKa = 6.76KK679 pKa = 11.05AEE681 pKa = 4.16VLYY684 pKa = 10.07GAPGTRR690 pKa = 11.84TVFEE694 pKa = 5.02LNWARR699 pKa = 11.84TRR701 pKa = 11.84KK702 pKa = 10.08GKK704 pKa = 10.55VGLRR708 pKa = 11.84LPVYY712 pKa = 8.96EE713 pKa = 4.52GSRR716 pKa = 11.84APVLAAEE723 pKa = 4.58MFRR726 pKa = 11.84YY727 pKa = 8.0TRR729 pKa = 11.84VGLARR734 pKa = 11.84VSEE737 pKa = 4.02EE738 pKa = 3.92LYY740 pKa = 10.65LRR742 pKa = 11.84EE743 pKa = 4.18SGIVPPDD750 pKa = 3.4VRR752 pKa = 11.84RR753 pKa = 11.84GRR755 pKa = 11.84AWKK758 pKa = 10.27CGLAVAILALGAALGYY774 pKa = 8.38TVYY777 pKa = 10.78ARR779 pKa = 11.84PPPKK783 pKa = 10.28PPDD786 pKa = 3.55PSVTVRR792 pKa = 11.84TGGTSS797 pKa = 2.79
MM1 pKa = 6.75QASRR5 pKa = 11.84MLEE8 pKa = 3.8SHH10 pKa = 6.39RR11 pKa = 11.84VWGEE15 pKa = 3.59YY16 pKa = 7.87RR17 pKa = 11.84THH19 pKa = 7.16RR20 pKa = 11.84LLEE23 pKa = 4.22AHH25 pKa = 7.37DD26 pKa = 5.42GRR28 pKa = 11.84ATWLRR33 pKa = 11.84AWTKK37 pKa = 10.23SWAPRR42 pKa = 11.84QGVVEE47 pKa = 4.18STPEE51 pKa = 3.78AEE53 pKa = 4.01KK54 pKa = 10.7HH55 pKa = 5.5VGVMTYY61 pKa = 10.32VKK63 pKa = 9.87GWRR66 pKa = 11.84RR67 pKa = 11.84RR68 pKa = 11.84LRR70 pKa = 11.84RR71 pKa = 11.84QLRR74 pKa = 11.84SARR77 pKa = 11.84TQFEE81 pKa = 4.24RR82 pKa = 11.84EE83 pKa = 4.19VVHH86 pKa = 6.55GSWAALEE93 pKa = 4.18GLYY96 pKa = 10.49SEE98 pKa = 5.4ALEE101 pKa = 4.81HH102 pKa = 6.64SPTMDD107 pKa = 3.24LMYY110 pKa = 11.0GHH112 pKa = 6.75VLSLSVDD119 pKa = 3.49RR120 pKa = 11.84DD121 pKa = 3.41HH122 pKa = 7.5GFTRR126 pKa = 11.84SFPSWYY132 pKa = 9.64VSPAGFEE139 pKa = 3.78NVPIVKK145 pKa = 8.82RR146 pKa = 11.84TTAKK150 pKa = 10.9DD151 pKa = 3.1PFTRR155 pKa = 11.84MVEE158 pKa = 4.04TGPTGIVARR167 pKa = 11.84TMMKK171 pKa = 10.29VGGRR175 pKa = 11.84PQKK178 pKa = 10.03FTDD181 pKa = 3.43AAATSLAYY189 pKa = 9.41KK190 pKa = 9.21WLHH193 pKa = 5.59ATGLWSSVTDD203 pKa = 3.67ADD205 pKa = 4.04VVINVSASFDD215 pKa = 3.4TSRR218 pKa = 11.84VHH220 pKa = 5.78MEE222 pKa = 4.03KK223 pKa = 9.68FTAPRR228 pKa = 11.84PPTEE232 pKa = 4.29GTLDD236 pKa = 3.55MCWEE240 pKa = 4.21VVARR244 pKa = 11.84NFKK247 pKa = 9.76EE248 pKa = 4.0LGMTGLPWITVADD261 pKa = 4.65VIAPHH266 pKa = 6.76INGDD270 pKa = 3.7ACPGLLTSRR279 pKa = 11.84IYY281 pKa = 9.73RR282 pKa = 11.84TRR284 pKa = 11.84AQASSCTTSDD294 pKa = 3.21VVALFEE300 pKa = 4.37ALRR303 pKa = 11.84TQEE306 pKa = 4.73LLCALPWAAGGRR318 pKa = 11.84EE319 pKa = 3.79KK320 pKa = 10.69RR321 pKa = 11.84QEE323 pKa = 4.12RR324 pKa = 11.84LVGDD328 pKa = 3.72QVSSRR333 pKa = 11.84LVQMPEE339 pKa = 3.76DD340 pKa = 3.38VLARR344 pKa = 11.84LEE346 pKa = 4.27SAVAGPLLQRR356 pKa = 11.84LKK358 pKa = 9.8WVKK361 pKa = 10.39RR362 pKa = 11.84DD363 pKa = 3.16IALGMSFFRR372 pKa = 11.84GRR374 pKa = 11.84AQRR377 pKa = 11.84FFHH380 pKa = 7.19RR381 pKa = 11.84FDD383 pKa = 3.17TCTHH387 pKa = 5.6VKK389 pKa = 10.49SFDD392 pKa = 2.93WSAFDD397 pKa = 3.61SSVGEE402 pKa = 4.12TLIVASFSLLRR413 pKa = 11.84ACYY416 pKa = 9.9PEE418 pKa = 4.72GQHH421 pKa = 6.6ADD423 pKa = 3.31NVFKK427 pKa = 11.13YY428 pKa = 9.68ILSAFVHH435 pKa = 5.85KK436 pKa = 10.29HH437 pKa = 5.92LIVPQGDD444 pKa = 4.06VIGMHH449 pKa = 6.74RR450 pKa = 11.84GIPSGSGLTSLVGSVANWLILRR472 pKa = 11.84TTLTDD477 pKa = 3.28ICPPLLAQRR486 pKa = 11.84AEE488 pKa = 4.22VAVLGDD494 pKa = 3.42DD495 pKa = 3.58TLIGFRR501 pKa = 11.84GYY503 pKa = 11.14DD504 pKa = 3.99GIPTNAEE511 pKa = 3.44FTARR515 pKa = 11.84AFALWGVAAGPSPSMVYY532 pKa = 10.35PEE534 pKa = 4.51GPPRR538 pKa = 11.84SVYY541 pKa = 10.5LEE543 pKa = 3.97RR544 pKa = 11.84CQTFLSVTSWCGLPSWRR561 pKa = 11.84NRR563 pKa = 11.84EE564 pKa = 3.66WWVMDD569 pKa = 3.04MGYY572 pKa = 11.08AKK574 pKa = 10.28GQPDD578 pKa = 3.47VANAQVAALEE588 pKa = 4.35DD589 pKa = 3.65AGFSGFAPQLRR600 pKa = 11.84GTRR603 pKa = 11.84RR604 pKa = 11.84RR605 pKa = 11.84FQKK608 pKa = 9.66WLQKK612 pKa = 10.53QPGAHH617 pKa = 6.2MLHH620 pKa = 6.83PEE622 pKa = 4.12YY623 pKa = 11.08VEE625 pKa = 4.02STEE628 pKa = 4.54TITQGWMARR637 pKa = 11.84AHH639 pKa = 6.41SSTGTAKK646 pKa = 10.35EE647 pKa = 3.69LAAFDD652 pKa = 4.07WDD654 pKa = 4.49SKK656 pKa = 10.39GQHH659 pKa = 6.38PGNWPTRR666 pKa = 11.84WKK668 pKa = 10.34VAAEE672 pKa = 3.99AAPPPHH678 pKa = 6.76KK679 pKa = 11.05AEE681 pKa = 4.16VLYY684 pKa = 10.07GAPGTRR690 pKa = 11.84TVFEE694 pKa = 5.02LNWARR699 pKa = 11.84TRR701 pKa = 11.84KK702 pKa = 10.08GKK704 pKa = 10.55VGLRR708 pKa = 11.84LPVYY712 pKa = 8.96EE713 pKa = 4.52GSRR716 pKa = 11.84APVLAAEE723 pKa = 4.58MFRR726 pKa = 11.84YY727 pKa = 8.0TRR729 pKa = 11.84VGLARR734 pKa = 11.84VSEE737 pKa = 4.02EE738 pKa = 3.92LYY740 pKa = 10.65LRR742 pKa = 11.84EE743 pKa = 4.18SGIVPPDD750 pKa = 3.4VRR752 pKa = 11.84RR753 pKa = 11.84GRR755 pKa = 11.84AWKK758 pKa = 10.27CGLAVAILALGAALGYY774 pKa = 8.38TVYY777 pKa = 10.78ARR779 pKa = 11.84PPPKK783 pKa = 10.28PPDD786 pKa = 3.55PSVTVRR792 pKa = 11.84TGGTSS797 pKa = 2.79
Molecular weight: 88.53 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1Z2RT46|A0A1Z2RT46_9VIRU RdRp OS=Leshenault partiti-like virus OX=2010278 PE=4 SV=1
MM1 pKa = 6.75QASRR5 pKa = 11.84MLEE8 pKa = 3.8SHH10 pKa = 6.39RR11 pKa = 11.84VWGEE15 pKa = 3.59YY16 pKa = 7.87RR17 pKa = 11.84THH19 pKa = 7.16RR20 pKa = 11.84LLEE23 pKa = 4.22AHH25 pKa = 7.37DD26 pKa = 5.42GRR28 pKa = 11.84ATWLRR33 pKa = 11.84AWTKK37 pKa = 10.23SWAPRR42 pKa = 11.84QGVVEE47 pKa = 4.18STPEE51 pKa = 3.78AEE53 pKa = 4.01KK54 pKa = 10.7HH55 pKa = 5.5VGVMTYY61 pKa = 10.32VKK63 pKa = 9.87GWRR66 pKa = 11.84RR67 pKa = 11.84RR68 pKa = 11.84LRR70 pKa = 11.84RR71 pKa = 11.84QLRR74 pKa = 11.84SARR77 pKa = 11.84TQFEE81 pKa = 4.24RR82 pKa = 11.84EE83 pKa = 4.19VVHH86 pKa = 6.55GSWAALEE93 pKa = 4.18GLYY96 pKa = 10.49SEE98 pKa = 5.4ALEE101 pKa = 4.81HH102 pKa = 6.64SPTMDD107 pKa = 3.24LMYY110 pKa = 11.0GHH112 pKa = 6.75VLSLSVDD119 pKa = 3.49RR120 pKa = 11.84DD121 pKa = 3.41HH122 pKa = 7.5GFTRR126 pKa = 11.84SFPSWYY132 pKa = 9.64VSPAGFEE139 pKa = 3.78NVPIVKK145 pKa = 8.82RR146 pKa = 11.84TTAKK150 pKa = 10.9DD151 pKa = 3.1PFTRR155 pKa = 11.84MVEE158 pKa = 4.04TGPTGIVARR167 pKa = 11.84TMMKK171 pKa = 10.29VGGRR175 pKa = 11.84PQKK178 pKa = 10.03FTDD181 pKa = 3.43AAATSLAYY189 pKa = 9.41KK190 pKa = 9.21WLHH193 pKa = 5.59ATGLWSSVTDD203 pKa = 3.67ADD205 pKa = 4.04VVINVSASFDD215 pKa = 3.4TSRR218 pKa = 11.84VHH220 pKa = 5.78MEE222 pKa = 4.03KK223 pKa = 9.68FTAPRR228 pKa = 11.84PPTEE232 pKa = 4.29GTLDD236 pKa = 3.55MCWEE240 pKa = 4.21VVARR244 pKa = 11.84NFKK247 pKa = 9.76EE248 pKa = 4.0LGMTGLPWITVADD261 pKa = 4.65VIAPHH266 pKa = 6.76INGDD270 pKa = 3.7ACPGLLTSRR279 pKa = 11.84IYY281 pKa = 9.73RR282 pKa = 11.84TRR284 pKa = 11.84AQASSCTTSDD294 pKa = 3.21VVALFEE300 pKa = 4.37ALRR303 pKa = 11.84TQEE306 pKa = 4.73LLCALPWAAGGRR318 pKa = 11.84EE319 pKa = 3.79KK320 pKa = 10.69RR321 pKa = 11.84QEE323 pKa = 4.12RR324 pKa = 11.84LVGDD328 pKa = 3.72QVSSRR333 pKa = 11.84LVQMPEE339 pKa = 3.76DD340 pKa = 3.38VLARR344 pKa = 11.84LEE346 pKa = 4.27SAVAGPLLQRR356 pKa = 11.84LKK358 pKa = 9.8WVKK361 pKa = 10.39RR362 pKa = 11.84DD363 pKa = 3.16IALGMSFFRR372 pKa = 11.84GRR374 pKa = 11.84AQRR377 pKa = 11.84FFHH380 pKa = 7.19RR381 pKa = 11.84FDD383 pKa = 3.17TCTHH387 pKa = 5.6VKK389 pKa = 10.49SFDD392 pKa = 2.93WSAFDD397 pKa = 3.61SSVGEE402 pKa = 4.12TLIVASFSLLRR413 pKa = 11.84ACYY416 pKa = 9.9PEE418 pKa = 4.72GQHH421 pKa = 6.6ADD423 pKa = 3.31NVFKK427 pKa = 11.13YY428 pKa = 9.68ILSAFVHH435 pKa = 5.85KK436 pKa = 10.29HH437 pKa = 5.92LIVPQGDD444 pKa = 4.06VIGMHH449 pKa = 6.74RR450 pKa = 11.84GIPSGSGLTSLVGSVANWLILRR472 pKa = 11.84TTLTDD477 pKa = 3.28ICPPLLAQRR486 pKa = 11.84AEE488 pKa = 4.22VAVLGDD494 pKa = 3.42DD495 pKa = 3.58TLIGFRR501 pKa = 11.84GYY503 pKa = 11.14DD504 pKa = 3.99GIPTNAEE511 pKa = 3.44FTARR515 pKa = 11.84AFALWGVAAGPSPSMVYY532 pKa = 10.35PEE534 pKa = 4.51GPPRR538 pKa = 11.84SVYY541 pKa = 10.5LEE543 pKa = 3.97RR544 pKa = 11.84CQTFLSVTSWCGLPSWRR561 pKa = 11.84NRR563 pKa = 11.84EE564 pKa = 3.66WWVMDD569 pKa = 3.04MGYY572 pKa = 11.08AKK574 pKa = 10.28GQPDD578 pKa = 3.47VANAQVAALEE588 pKa = 4.35DD589 pKa = 3.65AGFSGFAPQLRR600 pKa = 11.84GTRR603 pKa = 11.84RR604 pKa = 11.84RR605 pKa = 11.84FQKK608 pKa = 9.66WLQKK612 pKa = 10.53QPGAHH617 pKa = 6.2MLHH620 pKa = 6.83PEE622 pKa = 4.12YY623 pKa = 11.08VEE625 pKa = 4.02STEE628 pKa = 4.54TITQGWMARR637 pKa = 11.84AHH639 pKa = 6.41SSTGTAKK646 pKa = 10.35EE647 pKa = 3.69LAAFDD652 pKa = 4.07WDD654 pKa = 4.49SKK656 pKa = 10.39GQHH659 pKa = 6.38PGNWPTRR666 pKa = 11.84WKK668 pKa = 10.34VAAEE672 pKa = 3.99AAPPPHH678 pKa = 6.76KK679 pKa = 11.05AEE681 pKa = 4.16VLYY684 pKa = 10.07GAPGTRR690 pKa = 11.84TVFEE694 pKa = 5.02LNWARR699 pKa = 11.84TRR701 pKa = 11.84KK702 pKa = 10.08GKK704 pKa = 10.55VGLRR708 pKa = 11.84LPVYY712 pKa = 8.96EE713 pKa = 4.52GSRR716 pKa = 11.84APVLAAEE723 pKa = 4.58MFRR726 pKa = 11.84YY727 pKa = 8.0TRR729 pKa = 11.84VGLARR734 pKa = 11.84VSEE737 pKa = 4.02EE738 pKa = 3.92LYY740 pKa = 10.65LRR742 pKa = 11.84EE743 pKa = 4.18SGIVPPDD750 pKa = 3.4VRR752 pKa = 11.84RR753 pKa = 11.84GRR755 pKa = 11.84AWKK758 pKa = 10.27CGLAVAILALGAALGYY774 pKa = 8.38TVYY777 pKa = 10.78ARR779 pKa = 11.84PPPKK783 pKa = 10.28PPDD786 pKa = 3.55PSVTVRR792 pKa = 11.84TGGTSS797 pKa = 2.79
MM1 pKa = 6.75QASRR5 pKa = 11.84MLEE8 pKa = 3.8SHH10 pKa = 6.39RR11 pKa = 11.84VWGEE15 pKa = 3.59YY16 pKa = 7.87RR17 pKa = 11.84THH19 pKa = 7.16RR20 pKa = 11.84LLEE23 pKa = 4.22AHH25 pKa = 7.37DD26 pKa = 5.42GRR28 pKa = 11.84ATWLRR33 pKa = 11.84AWTKK37 pKa = 10.23SWAPRR42 pKa = 11.84QGVVEE47 pKa = 4.18STPEE51 pKa = 3.78AEE53 pKa = 4.01KK54 pKa = 10.7HH55 pKa = 5.5VGVMTYY61 pKa = 10.32VKK63 pKa = 9.87GWRR66 pKa = 11.84RR67 pKa = 11.84RR68 pKa = 11.84LRR70 pKa = 11.84RR71 pKa = 11.84QLRR74 pKa = 11.84SARR77 pKa = 11.84TQFEE81 pKa = 4.24RR82 pKa = 11.84EE83 pKa = 4.19VVHH86 pKa = 6.55GSWAALEE93 pKa = 4.18GLYY96 pKa = 10.49SEE98 pKa = 5.4ALEE101 pKa = 4.81HH102 pKa = 6.64SPTMDD107 pKa = 3.24LMYY110 pKa = 11.0GHH112 pKa = 6.75VLSLSVDD119 pKa = 3.49RR120 pKa = 11.84DD121 pKa = 3.41HH122 pKa = 7.5GFTRR126 pKa = 11.84SFPSWYY132 pKa = 9.64VSPAGFEE139 pKa = 3.78NVPIVKK145 pKa = 8.82RR146 pKa = 11.84TTAKK150 pKa = 10.9DD151 pKa = 3.1PFTRR155 pKa = 11.84MVEE158 pKa = 4.04TGPTGIVARR167 pKa = 11.84TMMKK171 pKa = 10.29VGGRR175 pKa = 11.84PQKK178 pKa = 10.03FTDD181 pKa = 3.43AAATSLAYY189 pKa = 9.41KK190 pKa = 9.21WLHH193 pKa = 5.59ATGLWSSVTDD203 pKa = 3.67ADD205 pKa = 4.04VVINVSASFDD215 pKa = 3.4TSRR218 pKa = 11.84VHH220 pKa = 5.78MEE222 pKa = 4.03KK223 pKa = 9.68FTAPRR228 pKa = 11.84PPTEE232 pKa = 4.29GTLDD236 pKa = 3.55MCWEE240 pKa = 4.21VVARR244 pKa = 11.84NFKK247 pKa = 9.76EE248 pKa = 4.0LGMTGLPWITVADD261 pKa = 4.65VIAPHH266 pKa = 6.76INGDD270 pKa = 3.7ACPGLLTSRR279 pKa = 11.84IYY281 pKa = 9.73RR282 pKa = 11.84TRR284 pKa = 11.84AQASSCTTSDD294 pKa = 3.21VVALFEE300 pKa = 4.37ALRR303 pKa = 11.84TQEE306 pKa = 4.73LLCALPWAAGGRR318 pKa = 11.84EE319 pKa = 3.79KK320 pKa = 10.69RR321 pKa = 11.84QEE323 pKa = 4.12RR324 pKa = 11.84LVGDD328 pKa = 3.72QVSSRR333 pKa = 11.84LVQMPEE339 pKa = 3.76DD340 pKa = 3.38VLARR344 pKa = 11.84LEE346 pKa = 4.27SAVAGPLLQRR356 pKa = 11.84LKK358 pKa = 9.8WVKK361 pKa = 10.39RR362 pKa = 11.84DD363 pKa = 3.16IALGMSFFRR372 pKa = 11.84GRR374 pKa = 11.84AQRR377 pKa = 11.84FFHH380 pKa = 7.19RR381 pKa = 11.84FDD383 pKa = 3.17TCTHH387 pKa = 5.6VKK389 pKa = 10.49SFDD392 pKa = 2.93WSAFDD397 pKa = 3.61SSVGEE402 pKa = 4.12TLIVASFSLLRR413 pKa = 11.84ACYY416 pKa = 9.9PEE418 pKa = 4.72GQHH421 pKa = 6.6ADD423 pKa = 3.31NVFKK427 pKa = 11.13YY428 pKa = 9.68ILSAFVHH435 pKa = 5.85KK436 pKa = 10.29HH437 pKa = 5.92LIVPQGDD444 pKa = 4.06VIGMHH449 pKa = 6.74RR450 pKa = 11.84GIPSGSGLTSLVGSVANWLILRR472 pKa = 11.84TTLTDD477 pKa = 3.28ICPPLLAQRR486 pKa = 11.84AEE488 pKa = 4.22VAVLGDD494 pKa = 3.42DD495 pKa = 3.58TLIGFRR501 pKa = 11.84GYY503 pKa = 11.14DD504 pKa = 3.99GIPTNAEE511 pKa = 3.44FTARR515 pKa = 11.84AFALWGVAAGPSPSMVYY532 pKa = 10.35PEE534 pKa = 4.51GPPRR538 pKa = 11.84SVYY541 pKa = 10.5LEE543 pKa = 3.97RR544 pKa = 11.84CQTFLSVTSWCGLPSWRR561 pKa = 11.84NRR563 pKa = 11.84EE564 pKa = 3.66WWVMDD569 pKa = 3.04MGYY572 pKa = 11.08AKK574 pKa = 10.28GQPDD578 pKa = 3.47VANAQVAALEE588 pKa = 4.35DD589 pKa = 3.65AGFSGFAPQLRR600 pKa = 11.84GTRR603 pKa = 11.84RR604 pKa = 11.84RR605 pKa = 11.84FQKK608 pKa = 9.66WLQKK612 pKa = 10.53QPGAHH617 pKa = 6.2MLHH620 pKa = 6.83PEE622 pKa = 4.12YY623 pKa = 11.08VEE625 pKa = 4.02STEE628 pKa = 4.54TITQGWMARR637 pKa = 11.84AHH639 pKa = 6.41SSTGTAKK646 pKa = 10.35EE647 pKa = 3.69LAAFDD652 pKa = 4.07WDD654 pKa = 4.49SKK656 pKa = 10.39GQHH659 pKa = 6.38PGNWPTRR666 pKa = 11.84WKK668 pKa = 10.34VAAEE672 pKa = 3.99AAPPPHH678 pKa = 6.76KK679 pKa = 11.05AEE681 pKa = 4.16VLYY684 pKa = 10.07GAPGTRR690 pKa = 11.84TVFEE694 pKa = 5.02LNWARR699 pKa = 11.84TRR701 pKa = 11.84KK702 pKa = 10.08GKK704 pKa = 10.55VGLRR708 pKa = 11.84LPVYY712 pKa = 8.96EE713 pKa = 4.52GSRR716 pKa = 11.84APVLAAEE723 pKa = 4.58MFRR726 pKa = 11.84YY727 pKa = 8.0TRR729 pKa = 11.84VGLARR734 pKa = 11.84VSEE737 pKa = 4.02EE738 pKa = 3.92LYY740 pKa = 10.65LRR742 pKa = 11.84EE743 pKa = 4.18SGIVPPDD750 pKa = 3.4VRR752 pKa = 11.84RR753 pKa = 11.84GRR755 pKa = 11.84AWKK758 pKa = 10.27CGLAVAILALGAALGYY774 pKa = 8.38TVYY777 pKa = 10.78ARR779 pKa = 11.84PPPKK783 pKa = 10.28PPDD786 pKa = 3.55PSVTVRR792 pKa = 11.84TGGTSS797 pKa = 2.79
Molecular weight: 88.53 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
797 |
797 |
797 |
797.0 |
88.53 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.414 ± 0.0 | 1.255 ± 0.0 |
4.015 ± 0.0 | 5.395 ± 0.0 |
3.764 ± 0.0 | 8.156 ± 0.0 |
2.76 ± 0.0 | 2.509 ± 0.0 |
3.388 ± 0.0 | 8.532 ± 0.0 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.509 ± 0.0 | 1.38 ± 0.0 |
6.274 ± 0.0 | 3.011 ± 0.0 |
8.657 ± 0.0 | 6.775 ± 0.0 |
7.026 ± 0.0 | 8.281 ± 0.0 |
3.388 ± 0.0 | 2.509 ± 0.0 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
