
Capybara microvirus Cap3_SP_586
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 6.76
Get precalculated fractions of proteins
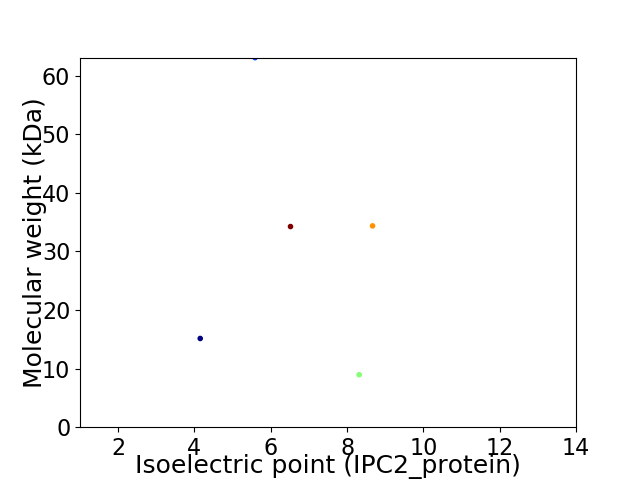
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
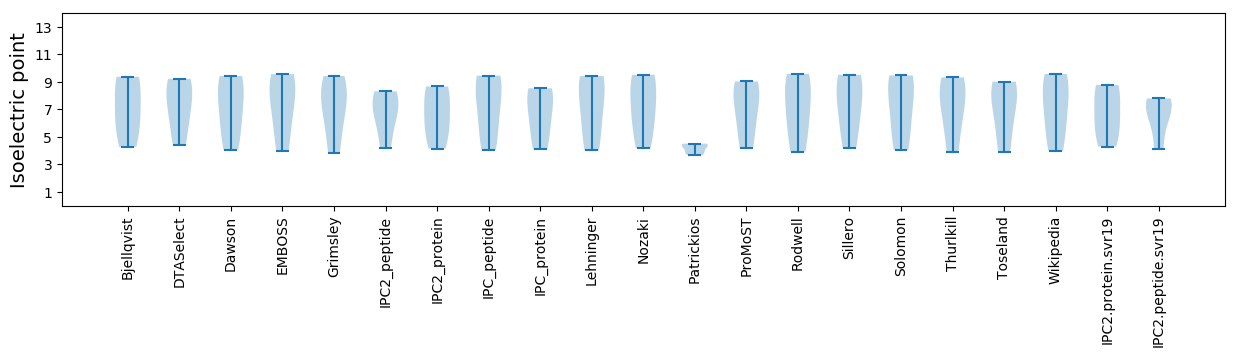
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4P8W8X8|A0A4P8W8X8_9VIRU Uncharacterized protein OS=Capybara microvirus Cap3_SP_586 OX=2585479 PE=4 SV=1
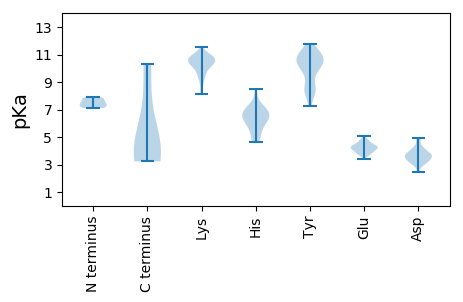
MM1 pKa = 7.32FRR3 pKa = 11.84NFCDD7 pKa = 3.3IILRR11 pKa = 11.84IMPSVSPQVFSSTSITQSGMDD32 pKa = 3.22VSLRR36 pKa = 11.84DD37 pKa = 3.83MISKK41 pKa = 9.8YY42 pKa = 7.26VTYY45 pKa = 10.14GVPIPDD51 pKa = 3.33VGGEE55 pKa = 4.2YY56 pKa = 10.81YY57 pKa = 11.03GDD59 pKa = 3.91DD60 pKa = 3.91TPDD63 pKa = 3.26VNPFGFDD70 pKa = 3.64LEE72 pKa = 4.88DD73 pKa = 3.7SPKK76 pKa = 10.06VRR78 pKa = 11.84QDD80 pKa = 3.45YY81 pKa = 11.16INAVNNAQALQNEE94 pKa = 4.56QTSQNASAMSPQNAQLAEE112 pKa = 4.17KK113 pKa = 10.67DD114 pKa = 3.73RR115 pKa = 11.84VNVASEE121 pKa = 4.08RR122 pKa = 11.84ASEE125 pKa = 3.99ASEE128 pKa = 3.82RR129 pKa = 11.84ASDD132 pKa = 3.16ITAPLEE138 pKa = 3.93
MM1 pKa = 7.32FRR3 pKa = 11.84NFCDD7 pKa = 3.3IILRR11 pKa = 11.84IMPSVSPQVFSSTSITQSGMDD32 pKa = 3.22VSLRR36 pKa = 11.84DD37 pKa = 3.83MISKK41 pKa = 9.8YY42 pKa = 7.26VTYY45 pKa = 10.14GVPIPDD51 pKa = 3.33VGGEE55 pKa = 4.2YY56 pKa = 10.81YY57 pKa = 11.03GDD59 pKa = 3.91DD60 pKa = 3.91TPDD63 pKa = 3.26VNPFGFDD70 pKa = 3.64LEE72 pKa = 4.88DD73 pKa = 3.7SPKK76 pKa = 10.06VRR78 pKa = 11.84QDD80 pKa = 3.45YY81 pKa = 11.16INAVNNAQALQNEE94 pKa = 4.56QTSQNASAMSPQNAQLAEE112 pKa = 4.17KK113 pKa = 10.67DD114 pKa = 3.73RR115 pKa = 11.84VNVASEE121 pKa = 4.08RR122 pKa = 11.84ASEE125 pKa = 3.99ASEE128 pKa = 3.82RR129 pKa = 11.84ASDD132 pKa = 3.16ITAPLEE138 pKa = 3.93
Molecular weight: 15.14 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4P8W555|A0A4P8W555_9VIRU Major capsid protein OS=Capybara microvirus Cap3_SP_586 OX=2585479 PE=3 SV=1
MM1 pKa = 7.11MCQSPIHH8 pKa = 6.42LRR10 pKa = 11.84DD11 pKa = 3.38GPLRR15 pKa = 11.84FVPCGKK21 pKa = 9.87CANCLKK27 pKa = 10.27RR28 pKa = 11.84KK29 pKa = 8.81HH30 pKa = 6.57LEE32 pKa = 3.6WQFRR36 pKa = 11.84LQRR39 pKa = 11.84EE40 pKa = 4.27EE41 pKa = 4.78RR42 pKa = 11.84SSKK45 pKa = 10.43CCYY48 pKa = 10.18CVTLTYY54 pKa = 10.77NEE56 pKa = 4.54SNVPISWTSKK66 pKa = 10.34NEE68 pKa = 3.69GGNLIFTLCKK78 pKa = 10.14SDD80 pKa = 3.27VQAWLKK86 pKa = 10.48RR87 pKa = 11.84IRR89 pKa = 11.84HH90 pKa = 5.3YY91 pKa = 10.85LPKK94 pKa = 10.04FRR96 pKa = 11.84YY97 pKa = 8.97YY98 pKa = 10.98LVGEE102 pKa = 4.48YY103 pKa = 8.36GTKK106 pKa = 8.32TLRR109 pKa = 11.84PHH111 pKa = 4.87YY112 pKa = 10.07HH113 pKa = 6.13VILFFSDD120 pKa = 3.63YY121 pKa = 11.17CNLQSVFDD129 pKa = 3.55ITFKK133 pKa = 10.71YY134 pKa = 8.17WCKK137 pKa = 10.87SDD139 pKa = 3.93IEE141 pKa = 4.25GHH143 pKa = 5.95KK144 pKa = 10.49VDD146 pKa = 4.19IVSPQAINYY155 pKa = 7.39CSKK158 pKa = 10.62YY159 pKa = 8.17VQKK162 pKa = 10.98LMSLPSWLDD171 pKa = 3.32CQPPFALMSRR181 pKa = 11.84KK182 pKa = 9.47PGIGDD187 pKa = 3.38SFIRR191 pKa = 11.84NEE193 pKa = 3.78ATATEE198 pKa = 4.11ILSNYY203 pKa = 8.58RR204 pKa = 11.84SQVFFDD210 pKa = 3.18EE211 pKa = 4.48GHH213 pKa = 6.38RR214 pKa = 11.84VTLPRR219 pKa = 11.84FYY221 pKa = 10.39RR222 pKa = 11.84DD223 pKa = 3.56KK224 pKa = 11.56LFIDD228 pKa = 4.07NLDD231 pKa = 3.39KK232 pKa = 11.03QIYY235 pKa = 8.91RR236 pKa = 11.84DD237 pKa = 3.09KK238 pKa = 10.82CKK240 pKa = 10.37RR241 pKa = 11.84YY242 pKa = 9.46FDD244 pKa = 4.61NYY246 pKa = 8.94TNQNFANYY254 pKa = 8.17VNKK257 pKa = 10.3FGFHH261 pKa = 4.88QACKK265 pKa = 10.02FFSDD269 pKa = 3.76MPKK272 pKa = 10.56NDD274 pKa = 3.57LQKK277 pKa = 10.96LIRR280 pKa = 11.84FFKK283 pKa = 10.76NKK285 pKa = 9.39QKK287 pKa = 11.05LL288 pKa = 3.49
MM1 pKa = 7.11MCQSPIHH8 pKa = 6.42LRR10 pKa = 11.84DD11 pKa = 3.38GPLRR15 pKa = 11.84FVPCGKK21 pKa = 9.87CANCLKK27 pKa = 10.27RR28 pKa = 11.84KK29 pKa = 8.81HH30 pKa = 6.57LEE32 pKa = 3.6WQFRR36 pKa = 11.84LQRR39 pKa = 11.84EE40 pKa = 4.27EE41 pKa = 4.78RR42 pKa = 11.84SSKK45 pKa = 10.43CCYY48 pKa = 10.18CVTLTYY54 pKa = 10.77NEE56 pKa = 4.54SNVPISWTSKK66 pKa = 10.34NEE68 pKa = 3.69GGNLIFTLCKK78 pKa = 10.14SDD80 pKa = 3.27VQAWLKK86 pKa = 10.48RR87 pKa = 11.84IRR89 pKa = 11.84HH90 pKa = 5.3YY91 pKa = 10.85LPKK94 pKa = 10.04FRR96 pKa = 11.84YY97 pKa = 8.97YY98 pKa = 10.98LVGEE102 pKa = 4.48YY103 pKa = 8.36GTKK106 pKa = 8.32TLRR109 pKa = 11.84PHH111 pKa = 4.87YY112 pKa = 10.07HH113 pKa = 6.13VILFFSDD120 pKa = 3.63YY121 pKa = 11.17CNLQSVFDD129 pKa = 3.55ITFKK133 pKa = 10.71YY134 pKa = 8.17WCKK137 pKa = 10.87SDD139 pKa = 3.93IEE141 pKa = 4.25GHH143 pKa = 5.95KK144 pKa = 10.49VDD146 pKa = 4.19IVSPQAINYY155 pKa = 7.39CSKK158 pKa = 10.62YY159 pKa = 8.17VQKK162 pKa = 10.98LMSLPSWLDD171 pKa = 3.32CQPPFALMSRR181 pKa = 11.84KK182 pKa = 9.47PGIGDD187 pKa = 3.38SFIRR191 pKa = 11.84NEE193 pKa = 3.78ATATEE198 pKa = 4.11ILSNYY203 pKa = 8.58RR204 pKa = 11.84SQVFFDD210 pKa = 3.18EE211 pKa = 4.48GHH213 pKa = 6.38RR214 pKa = 11.84VTLPRR219 pKa = 11.84FYY221 pKa = 10.39RR222 pKa = 11.84DD223 pKa = 3.56KK224 pKa = 11.56LFIDD228 pKa = 4.07NLDD231 pKa = 3.39KK232 pKa = 11.03QIYY235 pKa = 8.91RR236 pKa = 11.84DD237 pKa = 3.09KK238 pKa = 10.82CKK240 pKa = 10.37RR241 pKa = 11.84YY242 pKa = 9.46FDD244 pKa = 4.61NYY246 pKa = 8.94TNQNFANYY254 pKa = 8.17VNKK257 pKa = 10.3FGFHH261 pKa = 4.88QACKK265 pKa = 10.02FFSDD269 pKa = 3.76MPKK272 pKa = 10.56NDD274 pKa = 3.57LQKK277 pKa = 10.96LIRR280 pKa = 11.84FFKK283 pKa = 10.76NKK285 pKa = 9.39QKK287 pKa = 11.05LL288 pKa = 3.49
Molecular weight: 34.36 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1364 |
79 |
553 |
272.8 |
31.16 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.158 ± 1.379 | 1.613 ± 0.997 |
5.792 ± 0.554 | 4.619 ± 0.399 |
6.158 ± 0.975 | 5.425 ± 0.987 |
1.906 ± 0.336 | 5.865 ± 0.322 |
6.158 ± 0.938 | 9.238 ± 0.788 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.493 ± 0.515 | 6.965 ± 1.087 |
4.692 ± 0.513 | 5.059 ± 0.433 |
4.545 ± 0.629 | 7.625 ± 0.825 |
5.059 ± 0.647 | 4.692 ± 0.509 |
1.246 ± 0.209 | 4.692 ± 0.426 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
