
Rhizobiales bacterium GAS113
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; unclassified Hyphomicrobiales
Average proteome isoelectric point is 7.02
Get precalculated fractions of proteins
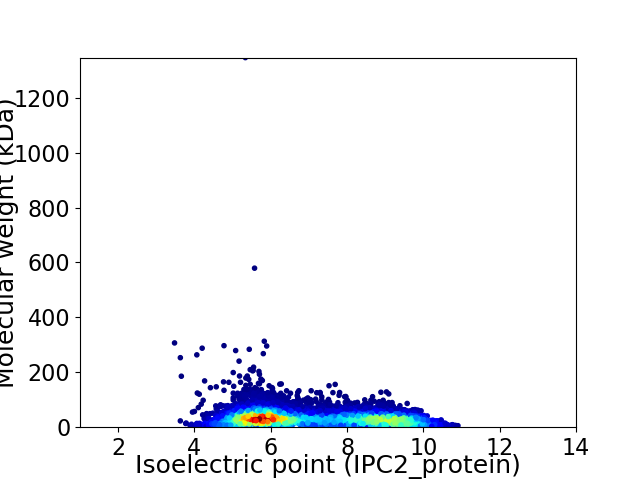
Virtual 2D-PAGE plot for 9038 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1H1ESL9|A0A1H1ESL9_9RHIZ Hydroxymethylpyrimidine kinase /phosphomethylpyrimidine kinase OS=Rhizobiales bacterium GAS113 OX=1884352 GN=SAMN05519103_00074 PE=4 SV=1
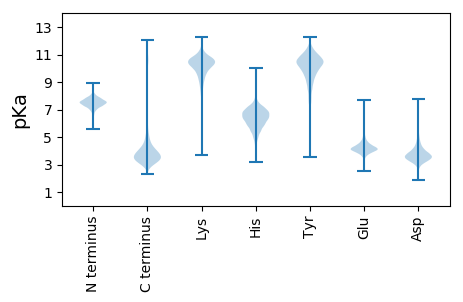
MM1 pKa = 7.84SDD3 pKa = 4.51PITNLHH9 pKa = 5.28YY10 pKa = 9.83TSGANLVNGQYY21 pKa = 10.56VPAADD26 pKa = 4.2GFNLADD32 pKa = 4.03VNSLDD37 pKa = 4.11QLNSLPSGVKK47 pKa = 10.07GLVYY51 pKa = 11.0LNMTGGATAAFQSAVSLYY69 pKa = 10.06IGNPKK74 pKa = 10.43LYY76 pKa = 10.52GFYY79 pKa = 10.6LVDD82 pKa = 4.16EE83 pKa = 5.32PDD85 pKa = 3.85PSLVPAANLKK95 pKa = 10.5SEE97 pKa = 4.63ADD99 pKa = 3.65WIHH102 pKa = 5.96AQLPGAITFTVLLNLGTPTNPTYY125 pKa = 11.33LNTYY129 pKa = 9.91NSANTDD135 pKa = 2.57IDD137 pKa = 5.01LFGLDD142 pKa = 4.68PYY144 pKa = 10.52PIRR147 pKa = 11.84PQFTNGVNYY156 pKa = 9.21TIIPAAVNAAEE167 pKa = 4.31AAGIAQSQIVPVYY180 pKa = 10.25QAFGDD185 pKa = 4.13SASYY189 pKa = 9.04TLPTASQEE197 pKa = 4.15QQILTTWGSVVPTPAFDD214 pKa = 3.91YY215 pKa = 10.38AYY217 pKa = 10.8SWGSQNGSLSLSGTPALQQIFAIHH241 pKa = 5.83NQPVSNTTVLNSVIEE256 pKa = 4.15APAVGISNIGNVVTITIGFSGTATVSGAPTLSLNDD291 pKa = 3.88NGTATYY297 pKa = 10.84VGGSGTNALVFSYY310 pKa = 10.29QVASSDD316 pKa = 3.22IDD318 pKa = 3.83VSSLAVTSVNLNGGTIHH335 pKa = 7.78DD336 pKa = 5.18AIGNNANLSLNAVTQGGPQIVTTVAKK362 pKa = 10.27DD363 pKa = 3.16QSLFSGISDD372 pKa = 3.7LTNYY376 pKa = 8.88PPHH379 pKa = 6.59NALAVGPNNIVMAEE393 pKa = 4.32GSRR396 pKa = 11.84IEE398 pKa = 4.14WTDD401 pKa = 3.27LTGGSATTQSVYY413 pKa = 11.35AFFGSLGTTATNSLFDD429 pKa = 3.67ARR431 pKa = 11.84AAYY434 pKa = 10.37DD435 pKa = 4.12SVNHH439 pKa = 6.52RR440 pKa = 11.84YY441 pKa = 8.96VVTMDD446 pKa = 4.09NIGSGGTISNVDD458 pKa = 3.02IAVSKK463 pKa = 10.7DD464 pKa = 3.33SNPADD469 pKa = 2.82GWYY472 pKa = 10.25FSSLNTSVTINGQLTASDD490 pKa = 3.97QPAVAVDD497 pKa = 3.51GTNIYY502 pKa = 8.53ITTPQYY508 pKa = 11.34NVNTSGWQGTEE519 pKa = 3.3LWVIGDD525 pKa = 3.61TAGAGGGIYY534 pKa = 10.35NGGTLTVAANQVALPGGSIFKK555 pKa = 10.34VAAGNNGKK563 pKa = 9.87SYY565 pKa = 10.95YY566 pKa = 10.53AGAYY570 pKa = 7.31STGDD574 pKa = 3.29HH575 pKa = 5.71TVVTVQTYY583 pKa = 11.2DD584 pKa = 3.05LGTNTFGTASTIALGNNDD602 pKa = 3.62QGNGGAEE609 pKa = 3.96LTAQQQGTNLLLDD622 pKa = 4.83AGDD625 pKa = 3.91STLQSLAYY633 pKa = 10.13AGGYY637 pKa = 10.16LYY639 pKa = 10.9GLSEE643 pKa = 4.4VKK645 pKa = 10.5PFGSGVPEE653 pKa = 3.84LHH655 pKa = 6.18WFKK658 pKa = 11.36LDD660 pKa = 3.38VSNPNNPIVVAQGDD674 pKa = 3.76VSGAAIGTNVATFDD688 pKa = 3.55GSIAVDD694 pKa = 3.71GAGDD698 pKa = 4.34AIINFTASGPNMYY711 pKa = 9.66PADD714 pKa = 3.75YY715 pKa = 10.26YY716 pKa = 10.91VYY718 pKa = 10.42HH719 pKa = 6.9GAADD723 pKa = 3.7PTGSFGAPVLYY734 pKa = 10.02QASTGFFNSGNGAGTQDD751 pKa = 2.5WGTRR755 pKa = 11.84SSAIADD761 pKa = 3.68PNNPHH766 pKa = 6.0SFWVSGEE773 pKa = 3.98YY774 pKa = 9.63VANGWWQTSAAQVAVQTVTAIIAPVVSSIAASGPGITGGNGDD816 pKa = 4.08LNAGKK821 pKa = 10.4VVTLTVNFSAPVIVNTTGGSPMLSLNDD848 pKa = 4.07GGSATYY854 pKa = 10.5AGGSGTSALTFSYY867 pKa = 10.01TVAAGQNTPDD877 pKa = 3.94LVVSALSLNGATIQGAAANNADD899 pKa = 4.52LSGATNSNPAGILQIDD915 pKa = 3.86TTSPVVTITSMGGSVNQATQTVTGTVTDD943 pKa = 4.1ADD945 pKa = 3.97GGTTGTTVTLFDD957 pKa = 3.63GTAQAGTATVQADD970 pKa = 4.28GSWSASVTLVNGTNTLTAKK989 pKa = 9.82DD990 pKa = 3.83TDD992 pKa = 3.93PAGNTGTSNGVTYY1005 pKa = 9.95TLTSPAPVVSSIATSGPGITGGNGDD1030 pKa = 4.08LNAGKK1035 pKa = 10.4VVTLTVNFSAPVTVNTTGGSPMLSLNDD1062 pKa = 4.02GGSASYY1068 pKa = 10.89AGGSGTSALTFSYY1081 pKa = 9.99TVAVGQNTPDD1091 pKa = 3.46LTVSALSLNGATIQDD1106 pKa = 3.56AAANNADD1113 pKa = 4.24LSGATNSNPAGILQIDD1129 pKa = 3.86TTSPVVTITSMGGSVNQATQTVTGTVTDD1157 pKa = 4.1ADD1159 pKa = 3.97GGTTGTTVTLFDD1171 pKa = 3.63GTAQAGTATVQADD1184 pKa = 4.13GSWNASVTLVNGTNTLTAKK1203 pKa = 9.82DD1204 pKa = 3.83TDD1206 pKa = 3.93PAGNTGTSNGVTYY1219 pKa = 9.95TLTSPAPVVSSIATSGPGISGGNGDD1244 pKa = 4.28LNAGKK1249 pKa = 10.4VVTLTVNFSAPVTVNTTGGSPTLSLNDD1276 pKa = 4.11GGSATYY1282 pKa = 10.5AGGSGTSALTFSYY1295 pKa = 10.01TVAAGQNSPDD1305 pKa = 3.62LTVSALTLNGATIQDD1320 pKa = 3.72AAATNADD1327 pKa = 3.69LSGATNSNPAGILQIDD1343 pKa = 3.86TTSPVVTITSMGGSVNQATQTLTGTVTDD1371 pKa = 4.47ADD1373 pKa = 3.97GGTTGTTVTLFDD1385 pKa = 3.63GTAQAGTATVQADD1398 pKa = 4.28GSWSASVTLVNGTNTLTAKK1417 pKa = 9.82DD1418 pKa = 3.83TDD1420 pKa = 3.93PAGNTGTSNGVTYY1433 pKa = 9.95TLTSPAPVVSSIATSGPGISGGNGDD1458 pKa = 4.28LNAGKK1463 pKa = 10.4VVTLTVNFSAPVTVNTTGGSPTLSLNDD1490 pKa = 4.11GGSATYY1496 pKa = 10.5AGGSGTSALTFSYY1509 pKa = 10.01TVAAGQNTPDD1519 pKa = 3.69LTVSALTLNGATIQDD1534 pKa = 3.72AAATNADD1541 pKa = 3.69LSGATNSNPAGILQIDD1557 pKa = 4.01TTSPTLAIATIASNNVINASTASAGFAISGTSTDD1591 pKa = 3.89AEE1593 pKa = 4.16NGQIVTVNIANSSNTVVDD1611 pKa = 4.39SYY1613 pKa = 9.54TTTDD1617 pKa = 3.3QGNAWSVNVTSAQATALADD1636 pKa = 3.29GRR1638 pKa = 11.84YY1639 pKa = 7.01TVTANVADD1647 pKa = 4.03QAGNAAPQASGALTVDD1663 pKa = 3.28EE1664 pKa = 5.42DD1665 pKa = 4.03KK1666 pKa = 11.31AAEE1669 pKa = 4.22APALTIANTALTVPAGGSVPLGITATQLDD1698 pKa = 3.99SDD1700 pKa = 4.51DD1701 pKa = 4.54RR1702 pKa = 11.84LTVTISGVPKK1712 pKa = 10.3YY1713 pKa = 10.66EE1714 pKa = 4.94SITAPSGDD1722 pKa = 3.48SVTSKK1727 pKa = 10.55RR1728 pKa = 11.84LSNGTYY1734 pKa = 8.61TWTITEE1740 pKa = 4.05NAGAAGTPLAGLMLKK1755 pKa = 10.27SSYY1758 pKa = 10.52AGSGHH1763 pKa = 7.41PIATFRR1769 pKa = 11.84VTASNATSGEE1779 pKa = 4.38TGTSTARR1786 pKa = 11.84SITVTDD1792 pKa = 4.63PPATTSATSLNSVAPDD1808 pKa = 3.88LDD1810 pKa = 3.64AAAATSPASVCRR1822 pKa = 11.84LAMVFNQFMAAGFQNDD1838 pKa = 3.42QNAAGQMTSLPQISSSLEE1856 pKa = 3.59DD1857 pKa = 3.33LAFLSRR1863 pKa = 11.84PHH1865 pKa = 6.66HH1866 pKa = 5.88SAA1868 pKa = 3.15
MM1 pKa = 7.84SDD3 pKa = 4.51PITNLHH9 pKa = 5.28YY10 pKa = 9.83TSGANLVNGQYY21 pKa = 10.56VPAADD26 pKa = 4.2GFNLADD32 pKa = 4.03VNSLDD37 pKa = 4.11QLNSLPSGVKK47 pKa = 10.07GLVYY51 pKa = 11.0LNMTGGATAAFQSAVSLYY69 pKa = 10.06IGNPKK74 pKa = 10.43LYY76 pKa = 10.52GFYY79 pKa = 10.6LVDD82 pKa = 4.16EE83 pKa = 5.32PDD85 pKa = 3.85PSLVPAANLKK95 pKa = 10.5SEE97 pKa = 4.63ADD99 pKa = 3.65WIHH102 pKa = 5.96AQLPGAITFTVLLNLGTPTNPTYY125 pKa = 11.33LNTYY129 pKa = 9.91NSANTDD135 pKa = 2.57IDD137 pKa = 5.01LFGLDD142 pKa = 4.68PYY144 pKa = 10.52PIRR147 pKa = 11.84PQFTNGVNYY156 pKa = 9.21TIIPAAVNAAEE167 pKa = 4.31AAGIAQSQIVPVYY180 pKa = 10.25QAFGDD185 pKa = 4.13SASYY189 pKa = 9.04TLPTASQEE197 pKa = 4.15QQILTTWGSVVPTPAFDD214 pKa = 3.91YY215 pKa = 10.38AYY217 pKa = 10.8SWGSQNGSLSLSGTPALQQIFAIHH241 pKa = 5.83NQPVSNTTVLNSVIEE256 pKa = 4.15APAVGISNIGNVVTITIGFSGTATVSGAPTLSLNDD291 pKa = 3.88NGTATYY297 pKa = 10.84VGGSGTNALVFSYY310 pKa = 10.29QVASSDD316 pKa = 3.22IDD318 pKa = 3.83VSSLAVTSVNLNGGTIHH335 pKa = 7.78DD336 pKa = 5.18AIGNNANLSLNAVTQGGPQIVTTVAKK362 pKa = 10.27DD363 pKa = 3.16QSLFSGISDD372 pKa = 3.7LTNYY376 pKa = 8.88PPHH379 pKa = 6.59NALAVGPNNIVMAEE393 pKa = 4.32GSRR396 pKa = 11.84IEE398 pKa = 4.14WTDD401 pKa = 3.27LTGGSATTQSVYY413 pKa = 11.35AFFGSLGTTATNSLFDD429 pKa = 3.67ARR431 pKa = 11.84AAYY434 pKa = 10.37DD435 pKa = 4.12SVNHH439 pKa = 6.52RR440 pKa = 11.84YY441 pKa = 8.96VVTMDD446 pKa = 4.09NIGSGGTISNVDD458 pKa = 3.02IAVSKK463 pKa = 10.7DD464 pKa = 3.33SNPADD469 pKa = 2.82GWYY472 pKa = 10.25FSSLNTSVTINGQLTASDD490 pKa = 3.97QPAVAVDD497 pKa = 3.51GTNIYY502 pKa = 8.53ITTPQYY508 pKa = 11.34NVNTSGWQGTEE519 pKa = 3.3LWVIGDD525 pKa = 3.61TAGAGGGIYY534 pKa = 10.35NGGTLTVAANQVALPGGSIFKK555 pKa = 10.34VAAGNNGKK563 pKa = 9.87SYY565 pKa = 10.95YY566 pKa = 10.53AGAYY570 pKa = 7.31STGDD574 pKa = 3.29HH575 pKa = 5.71TVVTVQTYY583 pKa = 11.2DD584 pKa = 3.05LGTNTFGTASTIALGNNDD602 pKa = 3.62QGNGGAEE609 pKa = 3.96LTAQQQGTNLLLDD622 pKa = 4.83AGDD625 pKa = 3.91STLQSLAYY633 pKa = 10.13AGGYY637 pKa = 10.16LYY639 pKa = 10.9GLSEE643 pKa = 4.4VKK645 pKa = 10.5PFGSGVPEE653 pKa = 3.84LHH655 pKa = 6.18WFKK658 pKa = 11.36LDD660 pKa = 3.38VSNPNNPIVVAQGDD674 pKa = 3.76VSGAAIGTNVATFDD688 pKa = 3.55GSIAVDD694 pKa = 3.71GAGDD698 pKa = 4.34AIINFTASGPNMYY711 pKa = 9.66PADD714 pKa = 3.75YY715 pKa = 10.26YY716 pKa = 10.91VYY718 pKa = 10.42HH719 pKa = 6.9GAADD723 pKa = 3.7PTGSFGAPVLYY734 pKa = 10.02QASTGFFNSGNGAGTQDD751 pKa = 2.5WGTRR755 pKa = 11.84SSAIADD761 pKa = 3.68PNNPHH766 pKa = 6.0SFWVSGEE773 pKa = 3.98YY774 pKa = 9.63VANGWWQTSAAQVAVQTVTAIIAPVVSSIAASGPGITGGNGDD816 pKa = 4.08LNAGKK821 pKa = 10.4VVTLTVNFSAPVIVNTTGGSPMLSLNDD848 pKa = 4.07GGSATYY854 pKa = 10.5AGGSGTSALTFSYY867 pKa = 10.01TVAAGQNTPDD877 pKa = 3.94LVVSALSLNGATIQGAAANNADD899 pKa = 4.52LSGATNSNPAGILQIDD915 pKa = 3.86TTSPVVTITSMGGSVNQATQTVTGTVTDD943 pKa = 4.1ADD945 pKa = 3.97GGTTGTTVTLFDD957 pKa = 3.63GTAQAGTATVQADD970 pKa = 4.28GSWSASVTLVNGTNTLTAKK989 pKa = 9.82DD990 pKa = 3.83TDD992 pKa = 3.93PAGNTGTSNGVTYY1005 pKa = 9.95TLTSPAPVVSSIATSGPGITGGNGDD1030 pKa = 4.08LNAGKK1035 pKa = 10.4VVTLTVNFSAPVTVNTTGGSPMLSLNDD1062 pKa = 4.02GGSASYY1068 pKa = 10.89AGGSGTSALTFSYY1081 pKa = 9.99TVAVGQNTPDD1091 pKa = 3.46LTVSALSLNGATIQDD1106 pKa = 3.56AAANNADD1113 pKa = 4.24LSGATNSNPAGILQIDD1129 pKa = 3.86TTSPVVTITSMGGSVNQATQTVTGTVTDD1157 pKa = 4.1ADD1159 pKa = 3.97GGTTGTTVTLFDD1171 pKa = 3.63GTAQAGTATVQADD1184 pKa = 4.13GSWNASVTLVNGTNTLTAKK1203 pKa = 9.82DD1204 pKa = 3.83TDD1206 pKa = 3.93PAGNTGTSNGVTYY1219 pKa = 9.95TLTSPAPVVSSIATSGPGISGGNGDD1244 pKa = 4.28LNAGKK1249 pKa = 10.4VVTLTVNFSAPVTVNTTGGSPTLSLNDD1276 pKa = 4.11GGSATYY1282 pKa = 10.5AGGSGTSALTFSYY1295 pKa = 10.01TVAAGQNSPDD1305 pKa = 3.62LTVSALTLNGATIQDD1320 pKa = 3.72AAATNADD1327 pKa = 3.69LSGATNSNPAGILQIDD1343 pKa = 3.86TTSPVVTITSMGGSVNQATQTLTGTVTDD1371 pKa = 4.47ADD1373 pKa = 3.97GGTTGTTVTLFDD1385 pKa = 3.63GTAQAGTATVQADD1398 pKa = 4.28GSWSASVTLVNGTNTLTAKK1417 pKa = 9.82DD1418 pKa = 3.83TDD1420 pKa = 3.93PAGNTGTSNGVTYY1433 pKa = 9.95TLTSPAPVVSSIATSGPGISGGNGDD1458 pKa = 4.28LNAGKK1463 pKa = 10.4VVTLTVNFSAPVTVNTTGGSPTLSLNDD1490 pKa = 4.11GGSATYY1496 pKa = 10.5AGGSGTSALTFSYY1509 pKa = 10.01TVAAGQNTPDD1519 pKa = 3.69LTVSALTLNGATIQDD1534 pKa = 3.72AAATNADD1541 pKa = 3.69LSGATNSNPAGILQIDD1557 pKa = 4.01TTSPTLAIATIASNNVINASTASAGFAISGTSTDD1591 pKa = 3.89AEE1593 pKa = 4.16NGQIVTVNIANSSNTVVDD1611 pKa = 4.39SYY1613 pKa = 9.54TTTDD1617 pKa = 3.3QGNAWSVNVTSAQATALADD1636 pKa = 3.29GRR1638 pKa = 11.84YY1639 pKa = 7.01TVTANVADD1647 pKa = 4.03QAGNAAPQASGALTVDD1663 pKa = 3.28EE1664 pKa = 5.42DD1665 pKa = 4.03KK1666 pKa = 11.31AAEE1669 pKa = 4.22APALTIANTALTVPAGGSVPLGITATQLDD1698 pKa = 3.99SDD1700 pKa = 4.51DD1701 pKa = 4.54RR1702 pKa = 11.84LTVTISGVPKK1712 pKa = 10.3YY1713 pKa = 10.66EE1714 pKa = 4.94SITAPSGDD1722 pKa = 3.48SVTSKK1727 pKa = 10.55RR1728 pKa = 11.84LSNGTYY1734 pKa = 8.61TWTITEE1740 pKa = 4.05NAGAAGTPLAGLMLKK1755 pKa = 10.27SSYY1758 pKa = 10.52AGSGHH1763 pKa = 7.41PIATFRR1769 pKa = 11.84VTASNATSGEE1779 pKa = 4.38TGTSTARR1786 pKa = 11.84SITVTDD1792 pKa = 4.63PPATTSATSLNSVAPDD1808 pKa = 3.88LDD1810 pKa = 3.64AAAATSPASVCRR1822 pKa = 11.84LAMVFNQFMAAGFQNDD1838 pKa = 3.42QNAAGQMTSLPQISSSLEE1856 pKa = 3.59DD1857 pKa = 3.33LAFLSRR1863 pKa = 11.84PHH1865 pKa = 6.66HH1866 pKa = 5.88SAA1868 pKa = 3.15
Molecular weight: 185.67 kDa
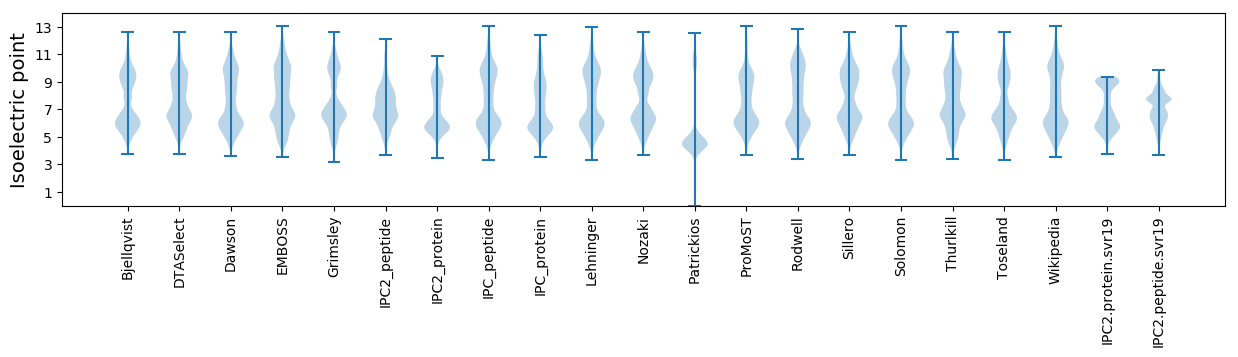
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1H1IAA7|A0A1H1IAA7_9RHIZ Uncharacterized protein OS=Rhizobiales bacterium GAS113 OX=1884352 GN=SAMN05519103_02209 PE=3 SV=1
MM1 pKa = 7.68GSGSLLISIATGAVAVVLVLGLINMLRR28 pKa = 11.84GGSPNRR34 pKa = 11.84SQNLMRR40 pKa = 11.84LRR42 pKa = 11.84VVLQAVAILLIVLVLWWRR60 pKa = 11.84GHH62 pKa = 4.79
MM1 pKa = 7.68GSGSLLISIATGAVAVVLVLGLINMLRR28 pKa = 11.84GGSPNRR34 pKa = 11.84SQNLMRR40 pKa = 11.84LRR42 pKa = 11.84VVLQAVAILLIVLVLWWRR60 pKa = 11.84GHH62 pKa = 4.79
Molecular weight: 6.62 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2831079 |
29 |
12478 |
313.2 |
33.89 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.059 ± 0.039 | 0.908 ± 0.009 |
5.26 ± 0.023 | 5.419 ± 0.028 |
3.709 ± 0.019 | 8.636 ± 0.035 |
2.06 ± 0.014 | 5.124 ± 0.018 |
3.135 ± 0.023 | 10.439 ± 0.037 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.306 ± 0.014 | 2.406 ± 0.019 |
5.375 ± 0.023 | 3.071 ± 0.016 |
7.531 ± 0.033 | 5.621 ± 0.022 |
5.171 ± 0.036 | 7.27 ± 0.02 |
1.337 ± 0.011 | 2.164 ± 0.012 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
