
Pneumovirus dog/Bari/100-12/ITA/2012
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Pneumoviridae; Orthopneumovirus; Murine orthopneumovirus
Average proteome isoelectric point is 6.77
Get precalculated fractions of proteins
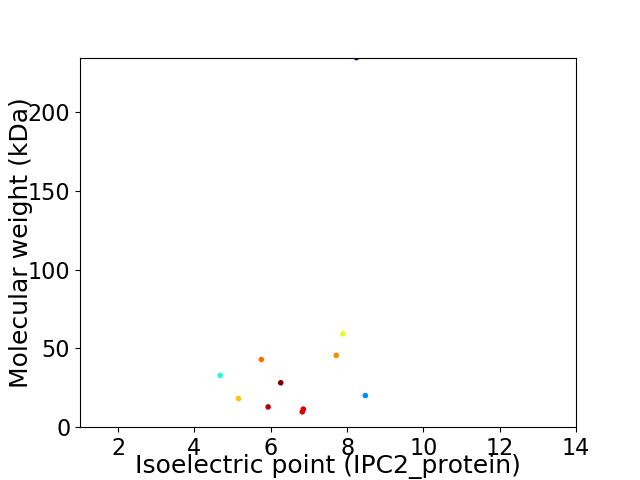
Virtual 2D-PAGE plot for 11 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|W0ISD5|W0ISD5_9MONO Protein M2-1 OS=Pneumovirus dog/Bari/100-12/ITA/2012 OX=2482952 GN=M2 PE=3 SV=1
MM1 pKa = 7.59EE2 pKa = 4.81KK3 pKa = 10.16FAPEE7 pKa = 4.1FVGEE11 pKa = 4.24DD12 pKa = 3.32ANKK15 pKa = 10.21KK16 pKa = 10.09AEE18 pKa = 4.2EE19 pKa = 4.03FLKK22 pKa = 10.78HH23 pKa = 6.57RR24 pKa = 11.84SFPSEE29 pKa = 4.14KK30 pKa = 9.72PLAGIPNTATHH41 pKa = 4.6VTKK44 pKa = 11.22YY45 pKa = 10.91NMPPILRR52 pKa = 11.84SSFKK56 pKa = 10.63LSPPRR61 pKa = 11.84AAAKK65 pKa = 8.23LTKK68 pKa = 9.98PSTPPSTPPPTPPQNKK84 pKa = 9.02EE85 pKa = 4.0EE86 pKa = 4.07QPKK89 pKa = 10.73EE90 pKa = 3.77SDD92 pKa = 3.12ADD94 pKa = 3.27IEE96 pKa = 4.56TIHH99 pKa = 7.75VYY101 pKa = 9.44EE102 pKa = 4.64VPDD105 pKa = 3.74NPEE108 pKa = 3.82HH109 pKa = 6.97SKK111 pKa = 10.94KK112 pKa = 10.2PCCSDD117 pKa = 3.33DD118 pKa = 3.54TDD120 pKa = 3.73TKK122 pKa = 10.05KK123 pKa = 8.88TRR125 pKa = 11.84KK126 pKa = 9.57PMVTFVEE133 pKa = 4.45PEE135 pKa = 3.94EE136 pKa = 4.52KK137 pKa = 10.32FVGLGASLYY146 pKa = 10.34KK147 pKa = 10.05EE148 pKa = 4.45TMQTFAADD156 pKa = 4.28GYY158 pKa = 10.96DD159 pKa = 3.57EE160 pKa = 4.49EE161 pKa = 5.77SNLSFEE167 pKa = 4.68EE168 pKa = 4.5TNQEE172 pKa = 4.04PGSSSVEE179 pKa = 3.72QRR181 pKa = 11.84LDD183 pKa = 3.74RR184 pKa = 11.84IEE186 pKa = 4.7EE187 pKa = 4.01KK188 pKa = 10.6LSYY191 pKa = 10.39IIGLLNTIMVATAGPTTARR210 pKa = 11.84DD211 pKa = 3.91EE212 pKa = 4.32IRR214 pKa = 11.84DD215 pKa = 3.73ALIGTRR221 pKa = 11.84EE222 pKa = 4.02EE223 pKa = 4.52LIEE226 pKa = 4.22MIKK229 pKa = 10.57SDD231 pKa = 3.56ILTVNDD237 pKa = 5.27RR238 pKa = 11.84IAAMEE243 pKa = 4.0KK244 pKa = 10.54LRR246 pKa = 11.84DD247 pKa = 3.97EE248 pKa = 4.57EE249 pKa = 4.34CSRR252 pKa = 11.84AEE254 pKa = 3.96TDD256 pKa = 5.15DD257 pKa = 5.08GSACYY262 pKa = 8.95LTDD265 pKa = 3.61RR266 pKa = 11.84ARR268 pKa = 11.84ILDD271 pKa = 4.28KK272 pKa = 10.67IVSSNAEE279 pKa = 3.88EE280 pKa = 4.12AKK282 pKa = 10.64EE283 pKa = 4.12DD284 pKa = 4.47LDD286 pKa = 3.84VDD288 pKa = 4.74DD289 pKa = 5.61IMGINFF295 pKa = 3.75
MM1 pKa = 7.59EE2 pKa = 4.81KK3 pKa = 10.16FAPEE7 pKa = 4.1FVGEE11 pKa = 4.24DD12 pKa = 3.32ANKK15 pKa = 10.21KK16 pKa = 10.09AEE18 pKa = 4.2EE19 pKa = 4.03FLKK22 pKa = 10.78HH23 pKa = 6.57RR24 pKa = 11.84SFPSEE29 pKa = 4.14KK30 pKa = 9.72PLAGIPNTATHH41 pKa = 4.6VTKK44 pKa = 11.22YY45 pKa = 10.91NMPPILRR52 pKa = 11.84SSFKK56 pKa = 10.63LSPPRR61 pKa = 11.84AAAKK65 pKa = 8.23LTKK68 pKa = 9.98PSTPPSTPPPTPPQNKK84 pKa = 9.02EE85 pKa = 4.0EE86 pKa = 4.07QPKK89 pKa = 10.73EE90 pKa = 3.77SDD92 pKa = 3.12ADD94 pKa = 3.27IEE96 pKa = 4.56TIHH99 pKa = 7.75VYY101 pKa = 9.44EE102 pKa = 4.64VPDD105 pKa = 3.74NPEE108 pKa = 3.82HH109 pKa = 6.97SKK111 pKa = 10.94KK112 pKa = 10.2PCCSDD117 pKa = 3.33DD118 pKa = 3.54TDD120 pKa = 3.73TKK122 pKa = 10.05KK123 pKa = 8.88TRR125 pKa = 11.84KK126 pKa = 9.57PMVTFVEE133 pKa = 4.45PEE135 pKa = 3.94EE136 pKa = 4.52KK137 pKa = 10.32FVGLGASLYY146 pKa = 10.34KK147 pKa = 10.05EE148 pKa = 4.45TMQTFAADD156 pKa = 4.28GYY158 pKa = 10.96DD159 pKa = 3.57EE160 pKa = 4.49EE161 pKa = 5.77SNLSFEE167 pKa = 4.68EE168 pKa = 4.5TNQEE172 pKa = 4.04PGSSSVEE179 pKa = 3.72QRR181 pKa = 11.84LDD183 pKa = 3.74RR184 pKa = 11.84IEE186 pKa = 4.7EE187 pKa = 4.01KK188 pKa = 10.6LSYY191 pKa = 10.39IIGLLNTIMVATAGPTTARR210 pKa = 11.84DD211 pKa = 3.91EE212 pKa = 4.32IRR214 pKa = 11.84DD215 pKa = 3.73ALIGTRR221 pKa = 11.84EE222 pKa = 4.02EE223 pKa = 4.52LIEE226 pKa = 4.22MIKK229 pKa = 10.57SDD231 pKa = 3.56ILTVNDD237 pKa = 5.27RR238 pKa = 11.84IAAMEE243 pKa = 4.0KK244 pKa = 10.54LRR246 pKa = 11.84DD247 pKa = 3.97EE248 pKa = 4.57EE249 pKa = 4.34CSRR252 pKa = 11.84AEE254 pKa = 3.96TDD256 pKa = 5.15DD257 pKa = 5.08GSACYY262 pKa = 8.95LTDD265 pKa = 3.61RR266 pKa = 11.84ARR268 pKa = 11.84ILDD271 pKa = 4.28KK272 pKa = 10.67IVSSNAEE279 pKa = 3.88EE280 pKa = 4.12AKK282 pKa = 10.64EE283 pKa = 4.12DD284 pKa = 4.47LDD286 pKa = 3.84VDD288 pKa = 4.74DD289 pKa = 5.61IMGINFF295 pKa = 3.75
Molecular weight: 32.89 kDa
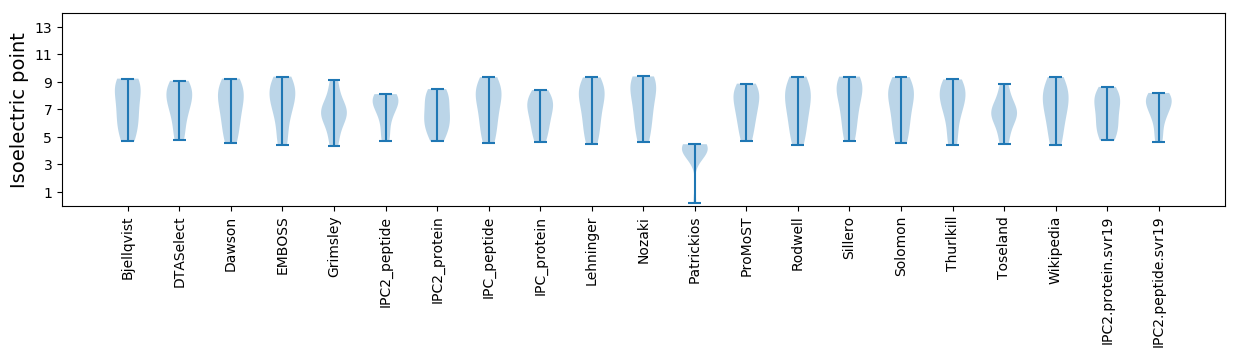
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|W0ISH6|W0ISH6_9MONO Non-structural protein 2 OS=Pneumovirus dog/Bari/100-12/ITA/2012 OX=2482952 GN=NS2 PE=4 SV=1
MM1 pKa = 7.5SVRR4 pKa = 11.84PCKK7 pKa = 10.54FEE9 pKa = 3.87VQGFCSRR16 pKa = 11.84GRR18 pKa = 11.84NCKK21 pKa = 9.77YY22 pKa = 9.6SHH24 pKa = 7.19KK25 pKa = 9.69YY26 pKa = 8.64WEE28 pKa = 4.56WPLKK32 pKa = 10.13TLMLRR37 pKa = 11.84QNYY40 pKa = 7.16MLNRR44 pKa = 11.84IYY46 pKa = 10.91RR47 pKa = 11.84FLDD50 pKa = 3.34TNTDD54 pKa = 3.0AMSDD58 pKa = 3.47VSGFDD63 pKa = 3.07APQRR67 pKa = 11.84TAEE70 pKa = 4.23YY71 pKa = 10.2ALGTIGVLKK80 pKa = 10.67SYY82 pKa = 10.88LEE84 pKa = 3.97KK85 pKa = 10.55TNNITKK91 pKa = 10.45SIACGSLITVLQNLDD106 pKa = 3.37VGLVIQARR114 pKa = 11.84DD115 pKa = 3.5SNAEE119 pKa = 3.72DD120 pKa = 3.37TNYY123 pKa = 10.27LRR125 pKa = 11.84SCNTILSYY133 pKa = 10.32IDD135 pKa = 4.64KK136 pKa = 9.89IHH138 pKa = 7.44KK139 pKa = 9.03KK140 pKa = 9.61RR141 pKa = 11.84QVIHH145 pKa = 6.62ILKK148 pKa = 9.24KK149 pKa = 10.24LPVGVLCSLIQSVISIEE166 pKa = 4.08EE167 pKa = 4.38KK168 pKa = 10.2INSSMKK174 pKa = 10.28TEE176 pKa = 3.83
MM1 pKa = 7.5SVRR4 pKa = 11.84PCKK7 pKa = 10.54FEE9 pKa = 3.87VQGFCSRR16 pKa = 11.84GRR18 pKa = 11.84NCKK21 pKa = 9.77YY22 pKa = 9.6SHH24 pKa = 7.19KK25 pKa = 9.69YY26 pKa = 8.64WEE28 pKa = 4.56WPLKK32 pKa = 10.13TLMLRR37 pKa = 11.84QNYY40 pKa = 7.16MLNRR44 pKa = 11.84IYY46 pKa = 10.91RR47 pKa = 11.84FLDD50 pKa = 3.34TNTDD54 pKa = 3.0AMSDD58 pKa = 3.47VSGFDD63 pKa = 3.07APQRR67 pKa = 11.84TAEE70 pKa = 4.23YY71 pKa = 10.2ALGTIGVLKK80 pKa = 10.67SYY82 pKa = 10.88LEE84 pKa = 3.97KK85 pKa = 10.55TNNITKK91 pKa = 10.45SIACGSLITVLQNLDD106 pKa = 3.37VGLVIQARR114 pKa = 11.84DD115 pKa = 3.5SNAEE119 pKa = 3.72DD120 pKa = 3.37TNYY123 pKa = 10.27LRR125 pKa = 11.84SCNTILSYY133 pKa = 10.32IDD135 pKa = 4.64KK136 pKa = 9.89IHH138 pKa = 7.44KK139 pKa = 9.03KK140 pKa = 9.61RR141 pKa = 11.84QVIHH145 pKa = 6.62ILKK148 pKa = 9.24KK149 pKa = 10.24LPVGVLCSLIQSVISIEE166 pKa = 4.08EE167 pKa = 4.38KK168 pKa = 10.2INSSMKK174 pKa = 10.28TEE176 pKa = 3.83
Molecular weight: 20.11 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
4571 |
92 |
2040 |
415.5 |
46.91 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.294 ± 1.031 | 2.231 ± 0.4 |
5.207 ± 0.427 | 5.163 ± 0.663 |
3.566 ± 0.319 | 4.9 ± 0.468 |
2.406 ± 0.311 | 6.913 ± 0.604 |
6.344 ± 0.47 | 10.785 ± 1.103 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.691 ± 0.174 | 5.907 ± 0.375 |
4.31 ± 0.805 | 3.128 ± 0.315 |
4.813 ± 0.231 | 8.335 ± 0.424 |
6.607 ± 0.758 | 6.454 ± 0.462 |
1.116 ± 0.236 | 3.807 ± 0.317 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
