
Pseudoalteromonas citrea DSM 8771
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Alteromonadales; Pseudoalteromonadaceae; Pseudoalteromonas; Pseudoalteromonas citrea
Average proteome isoelectric point is 6.27
Get precalculated fractions of proteins
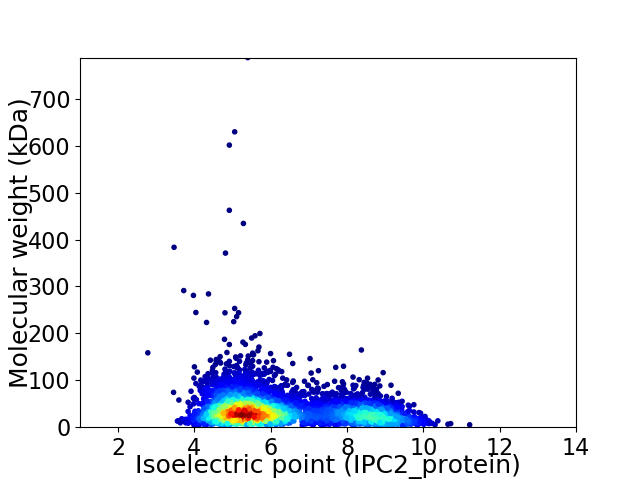
Virtual 2D-PAGE plot for 4457 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|U1KNS6|U1KNS6_9GAMM Polysaccharide export protein OS=Pseudoalteromonas citrea DSM 8771 OX=1117314 GN=PCIT_10764 PE=4 SV=1
MM1 pKa = 7.09TRR3 pKa = 11.84QRR5 pKa = 11.84LLGLILVSSSMLSGCGGGSDD25 pKa = 3.91SSSGTTPNTDD35 pKa = 3.03DD36 pKa = 3.92GGSVVVSNSAPVADD50 pKa = 4.85AGNEE54 pKa = 3.79QSVSVGTVVTLSGSQSQDD72 pKa = 2.56ADD74 pKa = 3.36GDD76 pKa = 4.11TLSFEE81 pKa = 4.32WQFITKK87 pKa = 8.96PASSQSSLVNATSSSTTFTPDD108 pKa = 2.3IAGQYY113 pKa = 8.73TVGLVVNDD121 pKa = 3.67GTVNSGQDD129 pKa = 3.25TVTVNAAAVNQMPIANAGSDD149 pKa = 3.59QQVLLGAVITLSGAASQDD167 pKa = 3.34PDD169 pKa = 4.16GDD171 pKa = 3.88TLSYY175 pKa = 10.66QWLFVSKK182 pKa = 10.59PNGSNAALTDD192 pKa = 3.75AQLVNAKK199 pKa = 8.6FTADD203 pKa = 3.06VAGNYY208 pKa = 8.86EE209 pKa = 4.19LGLTVSDD216 pKa = 4.09GQIEE220 pKa = 4.56STQDD224 pKa = 2.69TMTVVAQASTTNQAPIANAGNDD246 pKa = 3.53QTVSVADD253 pKa = 3.59VVTVSGALSSDD264 pKa = 3.24ADD266 pKa = 3.92NDD268 pKa = 4.17TLSYY272 pKa = 10.35AWSVVSKK279 pKa = 9.63PSQSNTVLSSSTSEE293 pKa = 4.3SVQLTPDD300 pKa = 3.16TAGTYY305 pKa = 9.3QLSLVVNDD313 pKa = 4.39GKK315 pKa = 11.51VNSAADD321 pKa = 4.09MITITAEE328 pKa = 4.0AQAGAPTANAGDD340 pKa = 4.11DD341 pKa = 3.56QSVTVGDD348 pKa = 4.97FVALSGSQSTGDD360 pKa = 3.35NLSYY364 pKa = 10.51QWQLVSKK371 pKa = 9.18PDD373 pKa = 3.41NSSAQLSAADD383 pKa = 3.57KK384 pKa = 11.15VSAEE388 pKa = 4.12FNADD392 pKa = 2.75IAGVFEE398 pKa = 4.31VSLTVTQGTSSATDD412 pKa = 3.41MLTVTSSNTSVNITDD427 pKa = 4.79KK428 pKa = 10.96IFTNEE433 pKa = 3.6AVSCRR438 pKa = 11.84NYY440 pKa = 10.03EE441 pKa = 3.55GSYY444 pKa = 10.17FSNVTDD450 pKa = 3.84IKK452 pKa = 10.61RR453 pKa = 11.84AADD456 pKa = 3.52FTGDD460 pKa = 3.28VVLTVGNTTCTITVNSIPNHH480 pKa = 6.38DD481 pKa = 4.5FNDD484 pKa = 3.66NTASFATNASAQNLVYY500 pKa = 10.35NLPITASFATATTSQGIGTTEE521 pKa = 3.82GVLLNGVTLDD531 pKa = 4.45LLPAACYY538 pKa = 10.22DD539 pKa = 4.0VGDD542 pKa = 4.01EE543 pKa = 4.41PIGRR547 pKa = 11.84EE548 pKa = 4.26KK549 pKa = 10.35IGCGPDD555 pKa = 3.4QNDD558 pKa = 3.36NPWRR562 pKa = 11.84YY563 pKa = 10.96DD564 pKa = 3.31PMSSLNTFGTDD575 pKa = 2.3AHH577 pKa = 6.36NAHH580 pKa = 6.12TQPSGKK586 pKa = 9.36YY587 pKa = 8.21HH588 pKa = 4.65YY589 pKa = 9.83HH590 pKa = 6.3GNPVAMFNQTCGSRR604 pKa = 11.84ASAVIGFAADD614 pKa = 4.0GFPIYY619 pKa = 10.52GSCFQDD625 pKa = 2.84ATTGTIRR632 pKa = 11.84KK633 pKa = 7.18ATSSYY638 pKa = 10.03VLKK641 pKa = 10.98NNGGARR647 pKa = 11.84QAVSGYY653 pKa = 4.85TTPVGGTGVVASNNYY668 pKa = 9.66DD669 pKa = 3.29GQFRR673 pKa = 11.84GDD675 pKa = 3.1WEE677 pKa = 4.41YY678 pKa = 11.08QASAGDD684 pKa = 3.79LDD686 pKa = 4.33EE687 pKa = 5.37CNGMTVNGQYY697 pKa = 10.93GYY699 pKa = 10.95YY700 pKa = 8.86VTDD703 pKa = 3.73TFPWVLGCFKK713 pKa = 11.17GNLNGTFTRR722 pKa = 11.84PTNLEE727 pKa = 4.05RR728 pKa = 11.84RR729 pKa = 11.84SHH731 pKa = 4.75SHH733 pKa = 5.78GSEE736 pKa = 3.58GSIAINGG743 pKa = 3.67
MM1 pKa = 7.09TRR3 pKa = 11.84QRR5 pKa = 11.84LLGLILVSSSMLSGCGGGSDD25 pKa = 3.91SSSGTTPNTDD35 pKa = 3.03DD36 pKa = 3.92GGSVVVSNSAPVADD50 pKa = 4.85AGNEE54 pKa = 3.79QSVSVGTVVTLSGSQSQDD72 pKa = 2.56ADD74 pKa = 3.36GDD76 pKa = 4.11TLSFEE81 pKa = 4.32WQFITKK87 pKa = 8.96PASSQSSLVNATSSSTTFTPDD108 pKa = 2.3IAGQYY113 pKa = 8.73TVGLVVNDD121 pKa = 3.67GTVNSGQDD129 pKa = 3.25TVTVNAAAVNQMPIANAGSDD149 pKa = 3.59QQVLLGAVITLSGAASQDD167 pKa = 3.34PDD169 pKa = 4.16GDD171 pKa = 3.88TLSYY175 pKa = 10.66QWLFVSKK182 pKa = 10.59PNGSNAALTDD192 pKa = 3.75AQLVNAKK199 pKa = 8.6FTADD203 pKa = 3.06VAGNYY208 pKa = 8.86EE209 pKa = 4.19LGLTVSDD216 pKa = 4.09GQIEE220 pKa = 4.56STQDD224 pKa = 2.69TMTVVAQASTTNQAPIANAGNDD246 pKa = 3.53QTVSVADD253 pKa = 3.59VVTVSGALSSDD264 pKa = 3.24ADD266 pKa = 3.92NDD268 pKa = 4.17TLSYY272 pKa = 10.35AWSVVSKK279 pKa = 9.63PSQSNTVLSSSTSEE293 pKa = 4.3SVQLTPDD300 pKa = 3.16TAGTYY305 pKa = 9.3QLSLVVNDD313 pKa = 4.39GKK315 pKa = 11.51VNSAADD321 pKa = 4.09MITITAEE328 pKa = 4.0AQAGAPTANAGDD340 pKa = 4.11DD341 pKa = 3.56QSVTVGDD348 pKa = 4.97FVALSGSQSTGDD360 pKa = 3.35NLSYY364 pKa = 10.51QWQLVSKK371 pKa = 9.18PDD373 pKa = 3.41NSSAQLSAADD383 pKa = 3.57KK384 pKa = 11.15VSAEE388 pKa = 4.12FNADD392 pKa = 2.75IAGVFEE398 pKa = 4.31VSLTVTQGTSSATDD412 pKa = 3.41MLTVTSSNTSVNITDD427 pKa = 4.79KK428 pKa = 10.96IFTNEE433 pKa = 3.6AVSCRR438 pKa = 11.84NYY440 pKa = 10.03EE441 pKa = 3.55GSYY444 pKa = 10.17FSNVTDD450 pKa = 3.84IKK452 pKa = 10.61RR453 pKa = 11.84AADD456 pKa = 3.52FTGDD460 pKa = 3.28VVLTVGNTTCTITVNSIPNHH480 pKa = 6.38DD481 pKa = 4.5FNDD484 pKa = 3.66NTASFATNASAQNLVYY500 pKa = 10.35NLPITASFATATTSQGIGTTEE521 pKa = 3.82GVLLNGVTLDD531 pKa = 4.45LLPAACYY538 pKa = 10.22DD539 pKa = 4.0VGDD542 pKa = 4.01EE543 pKa = 4.41PIGRR547 pKa = 11.84EE548 pKa = 4.26KK549 pKa = 10.35IGCGPDD555 pKa = 3.4QNDD558 pKa = 3.36NPWRR562 pKa = 11.84YY563 pKa = 10.96DD564 pKa = 3.31PMSSLNTFGTDD575 pKa = 2.3AHH577 pKa = 6.36NAHH580 pKa = 6.12TQPSGKK586 pKa = 9.36YY587 pKa = 8.21HH588 pKa = 4.65YY589 pKa = 9.83HH590 pKa = 6.3GNPVAMFNQTCGSRR604 pKa = 11.84ASAVIGFAADD614 pKa = 4.0GFPIYY619 pKa = 10.52GSCFQDD625 pKa = 2.84ATTGTIRR632 pKa = 11.84KK633 pKa = 7.18ATSSYY638 pKa = 10.03VLKK641 pKa = 10.98NNGGARR647 pKa = 11.84QAVSGYY653 pKa = 4.85TTPVGGTGVVASNNYY668 pKa = 9.66DD669 pKa = 3.29GQFRR673 pKa = 11.84GDD675 pKa = 3.1WEE677 pKa = 4.41YY678 pKa = 11.08QASAGDD684 pKa = 3.79LDD686 pKa = 4.33EE687 pKa = 5.37CNGMTVNGQYY697 pKa = 10.93GYY699 pKa = 10.95YY700 pKa = 8.86VTDD703 pKa = 3.73TFPWVLGCFKK713 pKa = 11.17GNLNGTFTRR722 pKa = 11.84PTNLEE727 pKa = 4.05RR728 pKa = 11.84RR729 pKa = 11.84SHH731 pKa = 4.75SHH733 pKa = 5.78GSEE736 pKa = 3.58GSIAINGG743 pKa = 3.67
Molecular weight: 76.67 kDa
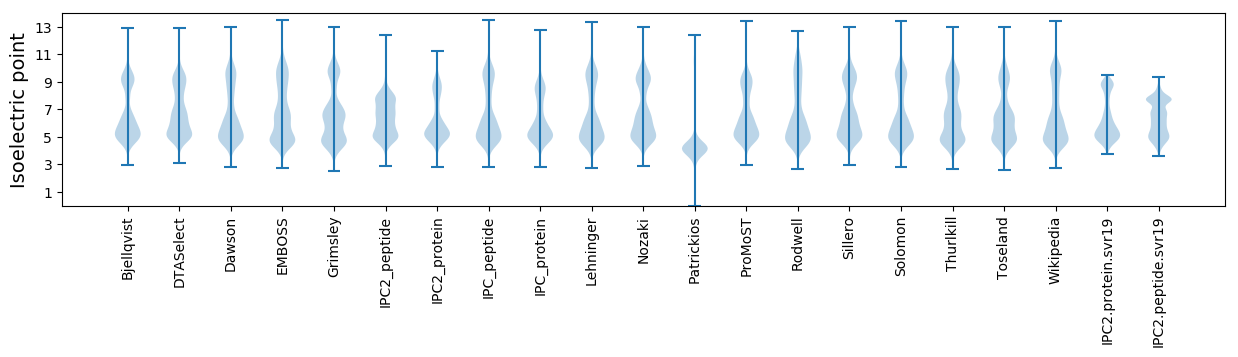
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|U1JJI4|U1JJI4_9GAMM Anti sigma E (Sigma 24) factor negative regulator OS=Pseudoalteromonas citrea DSM 8771 OX=1117314 GN=PCIT_18274 PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.42RR12 pKa = 11.84KK13 pKa = 9.04RR14 pKa = 11.84AHH16 pKa = 6.12GFRR19 pKa = 11.84ARR21 pKa = 11.84MATVNGRR28 pKa = 11.84KK29 pKa = 9.18VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.94GRR39 pKa = 11.84KK40 pKa = 8.67VLSAA44 pKa = 4.05
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.42RR12 pKa = 11.84KK13 pKa = 9.04RR14 pKa = 11.84AHH16 pKa = 6.12GFRR19 pKa = 11.84ARR21 pKa = 11.84MATVNGRR28 pKa = 11.84KK29 pKa = 9.18VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.94GRR39 pKa = 11.84KK40 pKa = 8.67VLSAA44 pKa = 4.05
Molecular weight: 5.09 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1556861 |
26 |
7131 |
349.3 |
38.88 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.441 ± 0.038 | 1.068 ± 0.012 |
5.55 ± 0.029 | 5.898 ± 0.039 |
4.193 ± 0.025 | 6.277 ± 0.041 |
2.515 ± 0.022 | 6.353 ± 0.028 |
5.547 ± 0.039 | 10.437 ± 0.046 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.288 ± 0.019 | 4.75 ± 0.027 |
3.663 ± 0.021 | 4.988 ± 0.034 |
4.049 ± 0.027 | 7.068 ± 0.037 |
5.694 ± 0.042 | 6.762 ± 0.033 |
1.157 ± 0.015 | 3.301 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
