
Spider associated circular virus 3
Taxonomy: Viruses; unclassified viruses; unclassified DNA viruses; unclassified ssDNA viruses
Average proteome isoelectric point is 7.56
Get precalculated fractions of proteins
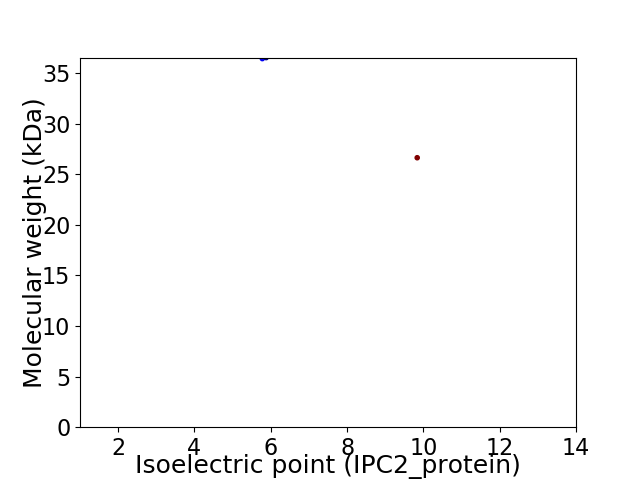
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A346BPE2|A0A346BPE2_9VIRU Putative capsid protein OS=Spider associated circular virus 3 OX=2293304 PE=4 SV=1
MM1 pKa = 7.36SRR3 pKa = 11.84TRR5 pKa = 11.84NWCFTIFTEE14 pKa = 5.48DD15 pKa = 3.37GDD17 pKa = 4.67GEE19 pKa = 4.32ASAHH23 pKa = 5.89YY24 pKa = 10.23DD25 pKa = 3.33PNTIDD30 pKa = 3.71WLSATPQVRR39 pKa = 11.84YY40 pKa = 9.92VVAGLEE46 pKa = 4.15VAPTSGRR53 pKa = 11.84LHH55 pKa = 5.05WQGYY59 pKa = 9.75IEE61 pKa = 4.5LQHH64 pKa = 6.69PATLVSCKK72 pKa = 10.39NSLGTDD78 pKa = 3.35KK79 pKa = 11.32AHH81 pKa = 7.11LEE83 pKa = 3.81IRR85 pKa = 11.84RR86 pKa = 11.84GTRR89 pKa = 11.84NQAIAYY95 pKa = 7.67CLKK98 pKa = 10.2QDD100 pKa = 3.7SAAQGEE106 pKa = 4.6PEE108 pKa = 4.22GNEE111 pKa = 3.67PKK113 pKa = 10.28IIFTWGDD120 pKa = 3.76DD121 pKa = 3.93NPQGTDD127 pKa = 3.47DD128 pKa = 4.64IYY130 pKa = 11.69DD131 pKa = 3.67EE132 pKa = 4.77VLCQTDD138 pKa = 3.74VNTAIQFIKK147 pKa = 10.24RR148 pKa = 11.84VRR150 pKa = 11.84PRR152 pKa = 11.84DD153 pKa = 3.43YY154 pKa = 10.97CIFGQQLHH162 pKa = 6.38TNLEE166 pKa = 4.02RR167 pKa = 11.84HH168 pKa = 4.47FTKK171 pKa = 10.53PKK173 pKa = 10.1AYY175 pKa = 10.19DD176 pKa = 3.29RR177 pKa = 11.84GTRR180 pKa = 11.84QYY182 pKa = 11.31NLPFLPDD189 pKa = 3.41HH190 pKa = 6.04VLEE193 pKa = 4.68TKK195 pKa = 10.42AIYY198 pKa = 10.3INGPAGSGKK207 pKa = 7.62TRR209 pKa = 11.84WSADD213 pKa = 3.0HH214 pKa = 6.97FEE216 pKa = 4.28NPLLVSHH223 pKa = 7.19KK224 pKa = 10.85DD225 pKa = 3.41DD226 pKa = 4.62LKK228 pKa = 11.03RR229 pKa = 11.84LAKK232 pKa = 10.31GSYY235 pKa = 9.85NGIIFDD241 pKa = 5.19DD242 pKa = 4.67FNISHH247 pKa = 6.76WPIGAVIHH255 pKa = 6.53LCDD258 pKa = 4.74LEE260 pKa = 4.15WDD262 pKa = 3.5RR263 pKa = 11.84SIDD266 pKa = 3.79VKK268 pKa = 10.91HH269 pKa = 5.67GTVTIPAQMPRR280 pKa = 11.84IFTNNLSFDD289 pKa = 3.65EE290 pKa = 4.48WCPSGLTEE298 pKa = 4.02EE299 pKa = 4.44QKK301 pKa = 10.42NALRR305 pKa = 11.84RR306 pKa = 11.84RR307 pKa = 11.84IHH309 pKa = 6.32VINVHH314 pKa = 4.93VRR316 pKa = 11.84LFEE319 pKa = 3.96
MM1 pKa = 7.36SRR3 pKa = 11.84TRR5 pKa = 11.84NWCFTIFTEE14 pKa = 5.48DD15 pKa = 3.37GDD17 pKa = 4.67GEE19 pKa = 4.32ASAHH23 pKa = 5.89YY24 pKa = 10.23DD25 pKa = 3.33PNTIDD30 pKa = 3.71WLSATPQVRR39 pKa = 11.84YY40 pKa = 9.92VVAGLEE46 pKa = 4.15VAPTSGRR53 pKa = 11.84LHH55 pKa = 5.05WQGYY59 pKa = 9.75IEE61 pKa = 4.5LQHH64 pKa = 6.69PATLVSCKK72 pKa = 10.39NSLGTDD78 pKa = 3.35KK79 pKa = 11.32AHH81 pKa = 7.11LEE83 pKa = 3.81IRR85 pKa = 11.84RR86 pKa = 11.84GTRR89 pKa = 11.84NQAIAYY95 pKa = 7.67CLKK98 pKa = 10.2QDD100 pKa = 3.7SAAQGEE106 pKa = 4.6PEE108 pKa = 4.22GNEE111 pKa = 3.67PKK113 pKa = 10.28IIFTWGDD120 pKa = 3.76DD121 pKa = 3.93NPQGTDD127 pKa = 3.47DD128 pKa = 4.64IYY130 pKa = 11.69DD131 pKa = 3.67EE132 pKa = 4.77VLCQTDD138 pKa = 3.74VNTAIQFIKK147 pKa = 10.24RR148 pKa = 11.84VRR150 pKa = 11.84PRR152 pKa = 11.84DD153 pKa = 3.43YY154 pKa = 10.97CIFGQQLHH162 pKa = 6.38TNLEE166 pKa = 4.02RR167 pKa = 11.84HH168 pKa = 4.47FTKK171 pKa = 10.53PKK173 pKa = 10.1AYY175 pKa = 10.19DD176 pKa = 3.29RR177 pKa = 11.84GTRR180 pKa = 11.84QYY182 pKa = 11.31NLPFLPDD189 pKa = 3.41HH190 pKa = 6.04VLEE193 pKa = 4.68TKK195 pKa = 10.42AIYY198 pKa = 10.3INGPAGSGKK207 pKa = 7.62TRR209 pKa = 11.84WSADD213 pKa = 3.0HH214 pKa = 6.97FEE216 pKa = 4.28NPLLVSHH223 pKa = 7.19KK224 pKa = 10.85DD225 pKa = 3.41DD226 pKa = 4.62LKK228 pKa = 11.03RR229 pKa = 11.84LAKK232 pKa = 10.31GSYY235 pKa = 9.85NGIIFDD241 pKa = 5.19DD242 pKa = 4.67FNISHH247 pKa = 6.76WPIGAVIHH255 pKa = 6.53LCDD258 pKa = 4.74LEE260 pKa = 4.15WDD262 pKa = 3.5RR263 pKa = 11.84SIDD266 pKa = 3.79VKK268 pKa = 10.91HH269 pKa = 5.67GTVTIPAQMPRR280 pKa = 11.84IFTNNLSFDD289 pKa = 3.65EE290 pKa = 4.48WCPSGLTEE298 pKa = 4.02EE299 pKa = 4.44QKK301 pKa = 10.42NALRR305 pKa = 11.84RR306 pKa = 11.84RR307 pKa = 11.84IHH309 pKa = 6.32VINVHH314 pKa = 4.93VRR316 pKa = 11.84LFEE319 pKa = 3.96
Molecular weight: 36.43 kDa
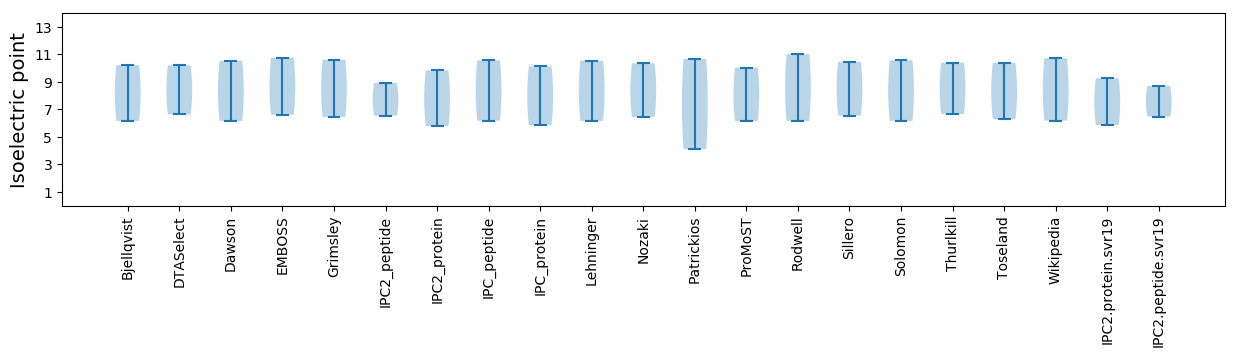
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A346BPE2|A0A346BPE2_9VIRU Putative capsid protein OS=Spider associated circular virus 3 OX=2293304 PE=4 SV=1
MM1 pKa = 7.69GYY3 pKa = 10.66GSKK6 pKa = 10.37FYY8 pKa = 10.75QKK10 pKa = 10.41RR11 pKa = 11.84KK12 pKa = 7.67YY13 pKa = 10.16QSPAYY18 pKa = 9.76FGLSPYY24 pKa = 9.84DD25 pKa = 3.3WKK27 pKa = 11.13RR28 pKa = 11.84PTSGYY33 pKa = 10.4YY34 pKa = 10.14YY35 pKa = 10.59GGFHH39 pKa = 6.68IRR41 pKa = 11.84RR42 pKa = 11.84RR43 pKa = 11.84VQRR46 pKa = 11.84GAINSNYY53 pKa = 9.58RR54 pKa = 11.84LKK56 pKa = 10.23STTKK60 pKa = 9.5SYY62 pKa = 10.48LRR64 pKa = 11.84KK65 pKa = 9.36RR66 pKa = 11.84GRR68 pKa = 11.84FVNHH72 pKa = 4.48TRR74 pKa = 11.84KK75 pKa = 10.46GKK77 pKa = 9.33FNRR80 pKa = 11.84YY81 pKa = 7.67HH82 pKa = 6.78HH83 pKa = 6.25ATSTGSRR90 pKa = 11.84RR91 pKa = 11.84FLAKK95 pKa = 10.34RR96 pKa = 11.84FPKK99 pKa = 10.15RR100 pKa = 11.84KK101 pKa = 9.29AVRR104 pKa = 11.84TITKK108 pKa = 10.14RR109 pKa = 11.84LVARR113 pKa = 11.84QKK115 pKa = 10.12PFPINKK121 pKa = 8.11EE122 pKa = 3.93ALGSDD127 pKa = 3.32IKK129 pKa = 9.06TTLLKK134 pKa = 10.42SVPAGTVVPLDD145 pKa = 4.48DD146 pKa = 4.56YY147 pKa = 11.26LIKK150 pKa = 11.11YY151 pKa = 9.71SDD153 pKa = 4.22LGVSSGLQHH162 pKa = 7.36DD163 pKa = 4.63NKK165 pKa = 10.3PLTWSFSTNLHH176 pKa = 4.58VKK178 pKa = 10.32YY179 pKa = 10.88YY180 pKa = 9.65LTFEE184 pKa = 4.2FAGACIDD191 pKa = 3.94LLVPQVPEE199 pKa = 3.74KK200 pKa = 11.18NIVKK204 pKa = 10.43KK205 pKa = 11.0LPDD208 pKa = 4.02ALNSITKK215 pKa = 9.91YY216 pKa = 11.02ADD218 pKa = 2.98VDD220 pKa = 3.91EE221 pKa = 5.36VDD223 pKa = 4.73TLMQTCNII231 pKa = 4.16
MM1 pKa = 7.69GYY3 pKa = 10.66GSKK6 pKa = 10.37FYY8 pKa = 10.75QKK10 pKa = 10.41RR11 pKa = 11.84KK12 pKa = 7.67YY13 pKa = 10.16QSPAYY18 pKa = 9.76FGLSPYY24 pKa = 9.84DD25 pKa = 3.3WKK27 pKa = 11.13RR28 pKa = 11.84PTSGYY33 pKa = 10.4YY34 pKa = 10.14YY35 pKa = 10.59GGFHH39 pKa = 6.68IRR41 pKa = 11.84RR42 pKa = 11.84RR43 pKa = 11.84VQRR46 pKa = 11.84GAINSNYY53 pKa = 9.58RR54 pKa = 11.84LKK56 pKa = 10.23STTKK60 pKa = 9.5SYY62 pKa = 10.48LRR64 pKa = 11.84KK65 pKa = 9.36RR66 pKa = 11.84GRR68 pKa = 11.84FVNHH72 pKa = 4.48TRR74 pKa = 11.84KK75 pKa = 10.46GKK77 pKa = 9.33FNRR80 pKa = 11.84YY81 pKa = 7.67HH82 pKa = 6.78HH83 pKa = 6.25ATSTGSRR90 pKa = 11.84RR91 pKa = 11.84FLAKK95 pKa = 10.34RR96 pKa = 11.84FPKK99 pKa = 10.15RR100 pKa = 11.84KK101 pKa = 9.29AVRR104 pKa = 11.84TITKK108 pKa = 10.14RR109 pKa = 11.84LVARR113 pKa = 11.84QKK115 pKa = 10.12PFPINKK121 pKa = 8.11EE122 pKa = 3.93ALGSDD127 pKa = 3.32IKK129 pKa = 9.06TTLLKK134 pKa = 10.42SVPAGTVVPLDD145 pKa = 4.48DD146 pKa = 4.56YY147 pKa = 11.26LIKK150 pKa = 11.11YY151 pKa = 9.71SDD153 pKa = 4.22LGVSSGLQHH162 pKa = 7.36DD163 pKa = 4.63NKK165 pKa = 10.3PLTWSFSTNLHH176 pKa = 4.58VKK178 pKa = 10.32YY179 pKa = 10.88YY180 pKa = 9.65LTFEE184 pKa = 4.2FAGACIDD191 pKa = 3.94LLVPQVPEE199 pKa = 3.74KK200 pKa = 11.18NIVKK204 pKa = 10.43KK205 pKa = 11.0LPDD208 pKa = 4.02ALNSITKK215 pKa = 9.91YY216 pKa = 11.02ADD218 pKa = 2.98VDD220 pKa = 3.91EE221 pKa = 5.36VDD223 pKa = 4.73TLMQTCNII231 pKa = 4.16
Molecular weight: 26.63 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
550 |
231 |
319 |
275.0 |
31.53 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.818 ± 0.337 | 1.636 ± 0.417 |
6.364 ± 0.867 | 3.818 ± 1.129 |
4.364 ± 0.216 | 6.545 ± 0.028 |
3.636 ± 0.562 | 6.0 ± 0.904 |
6.909 ± 1.883 | 8.0 ± 0.356 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
0.727 ± 0.075 | 4.909 ± 0.314 |
5.273 ± 0.042 | 3.818 ± 0.426 |
7.273 ± 0.515 | 5.818 ± 0.834 |
7.091 ± 0.145 | 5.455 ± 0.328 |
1.818 ± 0.515 | 4.727 ± 1.19 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
