
Streptococcus phage Javan330
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Siphoviridae; unclassified Siphoviridae
Average proteome isoelectric point is 6.37
Get precalculated fractions of proteins
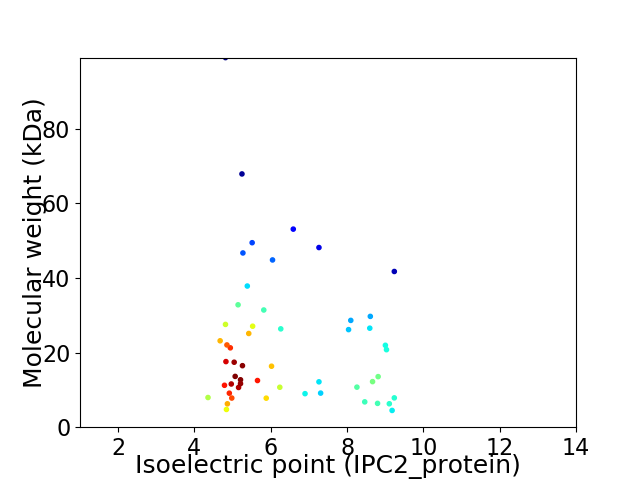
Virtual 2D-PAGE plot for 53 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4D6AZJ7|A0A4D6AZJ7_9CAUD Cro/CI family transcriptional regulator OS=Streptococcus phage Javan330 OX=2548110 GN=Javan330_0033 PE=4 SV=1
MM1 pKa = 7.74AEE3 pKa = 4.12YY4 pKa = 11.04VNQKK8 pKa = 8.82TGATINTNTEE18 pKa = 3.23ISGGDD23 pKa = 3.43WVPIAAYY30 pKa = 9.47KK31 pKa = 10.45PLDD34 pKa = 3.77SLTNAALKK42 pKa = 10.45EE43 pKa = 3.93ILDD46 pKa = 4.24EE47 pKa = 5.73KK48 pKa = 11.07GITYY52 pKa = 10.14DD53 pKa = 3.51NRR55 pKa = 11.84ATKK58 pKa = 10.34PEE60 pKa = 4.98LISLIEE66 pKa = 3.99QADD69 pKa = 3.97TEE71 pKa = 4.38AQQ73 pKa = 3.26
MM1 pKa = 7.74AEE3 pKa = 4.12YY4 pKa = 11.04VNQKK8 pKa = 8.82TGATINTNTEE18 pKa = 3.23ISGGDD23 pKa = 3.43WVPIAAYY30 pKa = 9.47KK31 pKa = 10.45PLDD34 pKa = 3.77SLTNAALKK42 pKa = 10.45EE43 pKa = 3.93ILDD46 pKa = 4.24EE47 pKa = 5.73KK48 pKa = 11.07GITYY52 pKa = 10.14DD53 pKa = 3.51NRR55 pKa = 11.84ATKK58 pKa = 10.34PEE60 pKa = 4.98LISLIEE66 pKa = 3.99QADD69 pKa = 3.97TEE71 pKa = 4.38AQQ73 pKa = 3.26
Molecular weight: 7.95 kDa
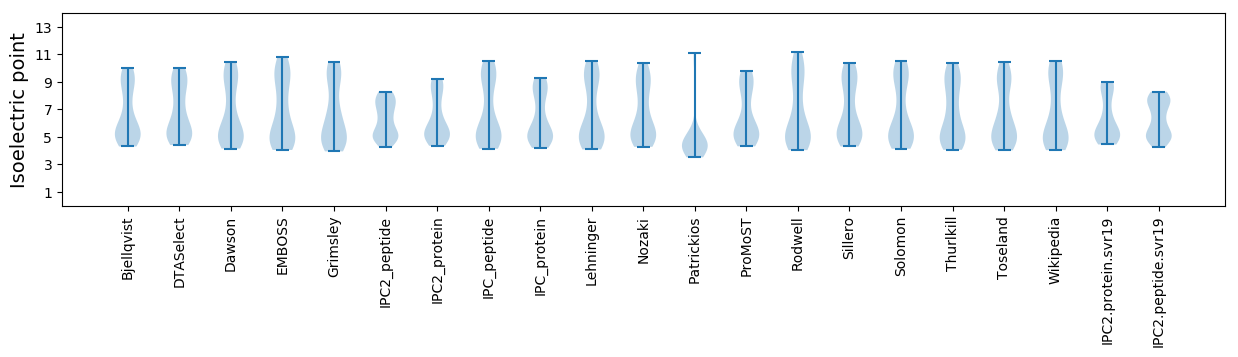
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4D6B152|A0A4D6B152_9CAUD Hyaluronidase OS=Streptococcus phage Javan330 OX=2548110 GN=Javan330_0009 PE=4 SV=1
MM1 pKa = 7.97EE2 pKa = 4.47IRR4 pKa = 11.84SYY6 pKa = 10.92KK7 pKa = 9.77KK8 pKa = 10.52KK9 pKa = 10.54NGEE12 pKa = 3.69TAYY15 pKa = 10.08MFRR18 pKa = 11.84AYY20 pKa = 9.41IGKK23 pKa = 9.64INGYY27 pKa = 7.31SQYY30 pKa = 9.8ATRR33 pKa = 11.84RR34 pKa = 11.84GFATKK39 pKa = 10.44AKK41 pKa = 10.21ARR43 pKa = 11.84AALLQLQDD51 pKa = 4.95DD52 pKa = 4.46IEE54 pKa = 5.14SGEE57 pKa = 3.99QSRR60 pKa = 11.84KK61 pKa = 9.67EE62 pKa = 3.66ITVEE66 pKa = 4.15EE67 pKa = 4.35IAKK70 pKa = 9.72KK71 pKa = 8.6WLKK74 pKa = 10.69DD75 pKa = 3.54YY76 pKa = 11.6SEE78 pKa = 4.59TVQDD82 pKa = 3.18STYY85 pKa = 10.82IKK87 pKa = 9.75TSRR90 pKa = 11.84NFKK93 pKa = 8.78NHH95 pKa = 6.67IYY97 pKa = 10.21PVFGNRR103 pKa = 11.84KK104 pKa = 7.76IASITPLQMQEE115 pKa = 4.15QANEE119 pKa = 3.72WSRR122 pKa = 11.84KK123 pKa = 8.08LVYY126 pKa = 9.94GRR128 pKa = 11.84KK129 pKa = 9.68LKK131 pKa = 11.2GLMNNVFKK139 pKa = 11.08YY140 pKa = 10.47AIRR143 pKa = 11.84HH144 pKa = 5.65GYY146 pKa = 9.54IDD148 pKa = 4.17TNPVDD153 pKa = 4.13SVIASTRR160 pKa = 11.84KK161 pKa = 10.06KK162 pKa = 10.77SDD164 pKa = 3.1GKK166 pKa = 11.05SDD168 pKa = 4.26FYY170 pKa = 11.86NKK172 pKa = 10.57DD173 pKa = 3.25EE174 pKa = 4.21LQKK177 pKa = 10.67FLKK180 pKa = 10.46LVSKK184 pKa = 9.77TKK186 pKa = 10.44DD187 pKa = 3.21LEE189 pKa = 4.81KK190 pKa = 10.02ITLFRR195 pKa = 11.84LLAFTGARR203 pKa = 11.84KK204 pKa = 10.34GEE206 pKa = 3.97ILALEE211 pKa = 4.27WNDD214 pKa = 3.14WTDD217 pKa = 2.97NTLDD221 pKa = 3.25INKK224 pKa = 10.21AITRR228 pKa = 11.84GFAGEE233 pKa = 4.32EE234 pKa = 3.65IGNTKK239 pKa = 8.91TVSSNRR245 pKa = 11.84LISLDD250 pKa = 3.71KK251 pKa = 9.58KK252 pKa = 9.13TKK254 pKa = 10.64NILKK258 pKa = 9.67KK259 pKa = 9.44WKK261 pKa = 9.47KK262 pKa = 9.65QNPNTKK268 pKa = 10.34YY269 pKa = 10.04IFEE272 pKa = 4.46NEE274 pKa = 4.01FKK276 pKa = 10.96KK277 pKa = 10.35PIPSTLPRR285 pKa = 11.84KK286 pKa = 8.65WLIKK290 pKa = 9.95IVEE293 pKa = 4.42GSDD296 pKa = 3.11LRR298 pKa = 11.84PIKK301 pKa = 10.35IHH303 pKa = 5.85GFRR306 pKa = 11.84HH307 pKa = 4.67THH309 pKa = 6.69ASLCFDD315 pKa = 4.04AGMTLKK321 pKa = 10.23QVQHH325 pKa = 6.86RR326 pKa = 11.84LGHH329 pKa = 6.18SDD331 pKa = 4.78LKK333 pKa = 9.01TTMNVYY339 pKa = 7.86THH341 pKa = 6.12ITKK344 pKa = 9.81QAKK347 pKa = 9.97DD348 pKa = 3.88DD349 pKa = 3.51IGEE352 pKa = 4.15RR353 pKa = 11.84FANYY357 pKa = 9.68IDD359 pKa = 3.83FF360 pKa = 4.89
MM1 pKa = 7.97EE2 pKa = 4.47IRR4 pKa = 11.84SYY6 pKa = 10.92KK7 pKa = 9.77KK8 pKa = 10.52KK9 pKa = 10.54NGEE12 pKa = 3.69TAYY15 pKa = 10.08MFRR18 pKa = 11.84AYY20 pKa = 9.41IGKK23 pKa = 9.64INGYY27 pKa = 7.31SQYY30 pKa = 9.8ATRR33 pKa = 11.84RR34 pKa = 11.84GFATKK39 pKa = 10.44AKK41 pKa = 10.21ARR43 pKa = 11.84AALLQLQDD51 pKa = 4.95DD52 pKa = 4.46IEE54 pKa = 5.14SGEE57 pKa = 3.99QSRR60 pKa = 11.84KK61 pKa = 9.67EE62 pKa = 3.66ITVEE66 pKa = 4.15EE67 pKa = 4.35IAKK70 pKa = 9.72KK71 pKa = 8.6WLKK74 pKa = 10.69DD75 pKa = 3.54YY76 pKa = 11.6SEE78 pKa = 4.59TVQDD82 pKa = 3.18STYY85 pKa = 10.82IKK87 pKa = 9.75TSRR90 pKa = 11.84NFKK93 pKa = 8.78NHH95 pKa = 6.67IYY97 pKa = 10.21PVFGNRR103 pKa = 11.84KK104 pKa = 7.76IASITPLQMQEE115 pKa = 4.15QANEE119 pKa = 3.72WSRR122 pKa = 11.84KK123 pKa = 8.08LVYY126 pKa = 9.94GRR128 pKa = 11.84KK129 pKa = 9.68LKK131 pKa = 11.2GLMNNVFKK139 pKa = 11.08YY140 pKa = 10.47AIRR143 pKa = 11.84HH144 pKa = 5.65GYY146 pKa = 9.54IDD148 pKa = 4.17TNPVDD153 pKa = 4.13SVIASTRR160 pKa = 11.84KK161 pKa = 10.06KK162 pKa = 10.77SDD164 pKa = 3.1GKK166 pKa = 11.05SDD168 pKa = 4.26FYY170 pKa = 11.86NKK172 pKa = 10.57DD173 pKa = 3.25EE174 pKa = 4.21LQKK177 pKa = 10.67FLKK180 pKa = 10.46LVSKK184 pKa = 9.77TKK186 pKa = 10.44DD187 pKa = 3.21LEE189 pKa = 4.81KK190 pKa = 10.02ITLFRR195 pKa = 11.84LLAFTGARR203 pKa = 11.84KK204 pKa = 10.34GEE206 pKa = 3.97ILALEE211 pKa = 4.27WNDD214 pKa = 3.14WTDD217 pKa = 2.97NTLDD221 pKa = 3.25INKK224 pKa = 10.21AITRR228 pKa = 11.84GFAGEE233 pKa = 4.32EE234 pKa = 3.65IGNTKK239 pKa = 8.91TVSSNRR245 pKa = 11.84LISLDD250 pKa = 3.71KK251 pKa = 9.58KK252 pKa = 9.13TKK254 pKa = 10.64NILKK258 pKa = 9.67KK259 pKa = 9.44WKK261 pKa = 9.47KK262 pKa = 9.65QNPNTKK268 pKa = 10.34YY269 pKa = 10.04IFEE272 pKa = 4.46NEE274 pKa = 4.01FKK276 pKa = 10.96KK277 pKa = 10.35PIPSTLPRR285 pKa = 11.84KK286 pKa = 8.65WLIKK290 pKa = 9.95IVEE293 pKa = 4.42GSDD296 pKa = 3.11LRR298 pKa = 11.84PIKK301 pKa = 10.35IHH303 pKa = 5.85GFRR306 pKa = 11.84HH307 pKa = 4.67THH309 pKa = 6.69ASLCFDD315 pKa = 4.04AGMTLKK321 pKa = 10.23QVQHH325 pKa = 6.86RR326 pKa = 11.84LGHH329 pKa = 6.18SDD331 pKa = 4.78LKK333 pKa = 9.01TTMNVYY339 pKa = 7.86THH341 pKa = 6.12ITKK344 pKa = 9.81QAKK347 pKa = 9.97DD348 pKa = 3.88DD349 pKa = 3.51IGEE352 pKa = 4.15RR353 pKa = 11.84FANYY357 pKa = 9.68IDD359 pKa = 3.83FF360 pKa = 4.89
Molecular weight: 41.73 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
10473 |
39 |
935 |
197.6 |
22.34 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.894 ± 0.519 | 0.582 ± 0.125 |
5.968 ± 0.208 | 7.639 ± 0.463 |
4.115 ± 0.263 | 6.913 ± 0.52 |
1.518 ± 0.182 | 6.894 ± 0.339 |
8.154 ± 0.452 | 8.107 ± 0.253 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.349 ± 0.139 | 5.481 ± 0.237 |
3.046 ± 0.272 | 3.838 ± 0.154 |
4.469 ± 0.282 | 6.388 ± 0.27 |
5.891 ± 0.282 | 6.245 ± 0.262 |
1.365 ± 0.158 | 4.144 ± 0.312 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
