
Capybara microvirus Cap3_SP_363
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 5.6
Get precalculated fractions of proteins
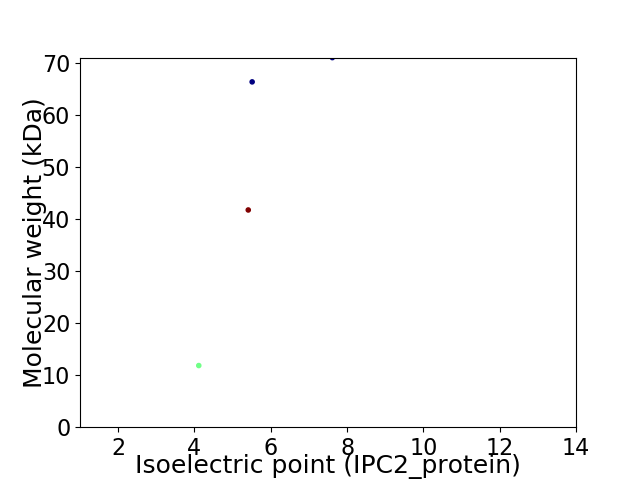
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
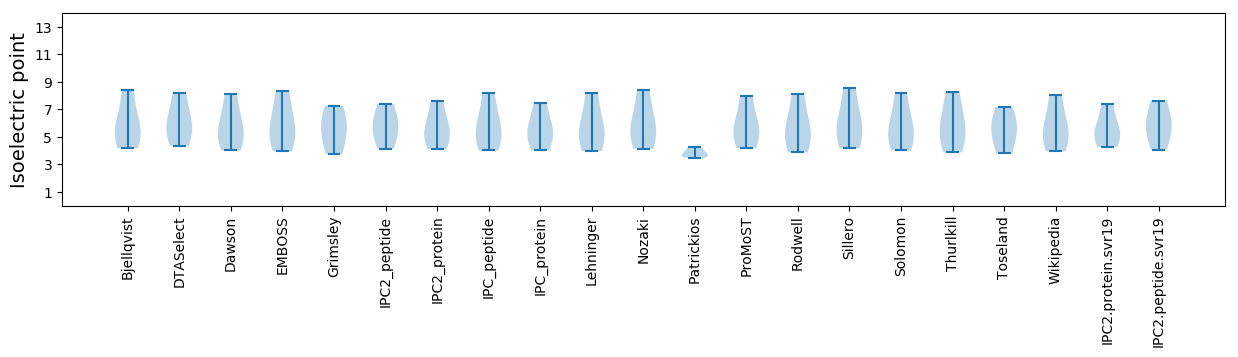
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4V1FVS5|A0A4V1FVS5_9VIRU Uncharacterized protein OS=Capybara microvirus Cap3_SP_363 OX=2585435 PE=4 SV=1
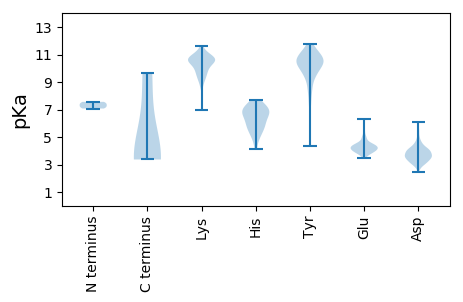
MM1 pKa = 7.07IRR3 pKa = 11.84KK4 pKa = 9.49LIGEE8 pKa = 4.41HH9 pKa = 5.87VPSVVQDD16 pKa = 4.52DD17 pKa = 3.47IRR19 pKa = 11.84LITVVDD25 pKa = 3.58EE26 pKa = 4.52EE27 pKa = 5.48GNVSSTFSKK36 pKa = 10.77YY37 pKa = 10.45DD38 pKa = 3.57PSSVSYY44 pKa = 10.87GCVGDD49 pKa = 3.57WSLNSLISAGINPSTLNLSTNNRR72 pKa = 11.84LNKK75 pKa = 9.71FDD77 pKa = 4.06EE78 pKa = 4.91FSDD81 pKa = 4.13FEE83 pKa = 5.15AYY85 pKa = 10.21ADD87 pKa = 3.73SIMRR91 pKa = 11.84DD92 pKa = 3.73VIIEE96 pKa = 4.19TPQVDD101 pKa = 3.98SNEE104 pKa = 4.16NNNN107 pKa = 3.37
MM1 pKa = 7.07IRR3 pKa = 11.84KK4 pKa = 9.49LIGEE8 pKa = 4.41HH9 pKa = 5.87VPSVVQDD16 pKa = 4.52DD17 pKa = 3.47IRR19 pKa = 11.84LITVVDD25 pKa = 3.58EE26 pKa = 4.52EE27 pKa = 5.48GNVSSTFSKK36 pKa = 10.77YY37 pKa = 10.45DD38 pKa = 3.57PSSVSYY44 pKa = 10.87GCVGDD49 pKa = 3.57WSLNSLISAGINPSTLNLSTNNRR72 pKa = 11.84LNKK75 pKa = 9.71FDD77 pKa = 4.06EE78 pKa = 4.91FSDD81 pKa = 4.13FEE83 pKa = 5.15AYY85 pKa = 10.21ADD87 pKa = 3.73SIMRR91 pKa = 11.84DD92 pKa = 3.73VIIEE96 pKa = 4.19TPQVDD101 pKa = 3.98SNEE104 pKa = 4.16NNNN107 pKa = 3.37
Molecular weight: 11.86 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4P8W5Q0|A0A4P8W5Q0_9VIRU Minor capsid protein OS=Capybara microvirus Cap3_SP_363 OX=2585435 PE=4 SV=1
MM1 pKa = 7.42LTQTLNIRR9 pKa = 11.84NNTHH13 pKa = 5.59YY14 pKa = 10.1FCPSGSRR21 pKa = 11.84LINSVINPSTEE32 pKa = 3.83RR33 pKa = 11.84YY34 pKa = 9.34KK35 pKa = 10.69SLRR38 pKa = 11.84VQGYY42 pKa = 6.98NQRR45 pKa = 11.84LYY47 pKa = 11.38YY48 pKa = 10.29EE49 pKa = 3.92YY50 pKa = 10.05TYY52 pKa = 10.59CKK54 pKa = 9.95SKK56 pKa = 10.67NYY58 pKa = 9.93PVFSYY63 pKa = 10.49TLTYY67 pKa = 10.59NNKK70 pKa = 9.25SLPMFYY76 pKa = 11.08GNAVHH81 pKa = 7.67DD82 pKa = 4.59YY83 pKa = 10.84NDD85 pKa = 2.45IRR87 pKa = 11.84YY88 pKa = 8.08FFRR91 pKa = 11.84EE92 pKa = 4.01SGFDD96 pKa = 3.23KK97 pKa = 10.87HH98 pKa = 6.69LLRR101 pKa = 11.84HH102 pKa = 5.64YY103 pKa = 10.69GSVVKK108 pKa = 9.98YY109 pKa = 8.81FVASEE114 pKa = 4.09LGEE117 pKa = 4.34GGISHH122 pKa = 6.8QDD124 pKa = 3.29SLSHH128 pKa = 5.82NNKK131 pKa = 9.16RR132 pKa = 11.84AQRR135 pKa = 11.84GKK137 pKa = 10.59DD138 pKa = 3.51NNPHH142 pKa = 4.87YY143 pKa = 10.7HH144 pKa = 6.35VIFFVYY150 pKa = 9.94NANDD154 pKa = 2.99TRR156 pKa = 11.84YY157 pKa = 9.71PYY159 pKa = 10.49IRR161 pKa = 11.84INGKK165 pKa = 6.96QFKK168 pKa = 9.89TIVQRR173 pKa = 11.84YY174 pKa = 4.38WQGDD178 pKa = 2.72AWRR181 pKa = 11.84TKK183 pKa = 10.33KK184 pKa = 9.52PQEE187 pKa = 4.04YY188 pKa = 10.19KK189 pKa = 10.84FGIAMNAKK197 pKa = 9.12YY198 pKa = 10.65NLGEE202 pKa = 4.1ITNGSAIRR210 pKa = 11.84YY211 pKa = 6.18NSKK214 pKa = 10.86YY215 pKa = 9.39VVKK218 pKa = 10.65DD219 pKa = 3.16VLQSRR224 pKa = 11.84KK225 pKa = 9.88DD226 pKa = 3.7DD227 pKa = 3.57SCLRR231 pKa = 11.84SCEE234 pKa = 4.36EE235 pKa = 3.8YY236 pKa = 9.75WNSVDD241 pKa = 3.45GEE243 pKa = 4.46YY244 pKa = 10.51FITHH248 pKa = 6.72HH249 pKa = 6.65KK250 pKa = 10.4LFWDD254 pKa = 4.44DD255 pKa = 3.57YY256 pKa = 10.06FDD258 pKa = 5.62RR259 pKa = 11.84YY260 pKa = 11.27VNDD263 pKa = 3.63FASDD267 pKa = 3.42VEE269 pKa = 4.6YY270 pKa = 10.74INGISDD276 pKa = 3.59VFGGFHH282 pKa = 6.62YY283 pKa = 10.5EE284 pKa = 3.94VADD287 pKa = 4.13FSGMSEE293 pKa = 4.02NEE295 pKa = 3.91LYY297 pKa = 10.61DD298 pKa = 3.9AYY300 pKa = 11.42VYY302 pKa = 10.32IYY304 pKa = 10.29GDD306 pKa = 3.17RR307 pKa = 11.84VNFDD311 pKa = 3.45NLLTEE316 pKa = 4.15HH317 pKa = 7.4RR318 pKa = 11.84DD319 pKa = 3.65YY320 pKa = 11.14IIDD323 pKa = 4.37LEE325 pKa = 4.16INRR328 pKa = 11.84YY329 pKa = 8.49KK330 pKa = 10.57NRR332 pKa = 11.84HH333 pKa = 4.14RR334 pKa = 11.84TKK336 pKa = 10.52VKK338 pKa = 9.07ISNGIGLSALDD349 pKa = 3.87KK350 pKa = 10.68MVDD353 pKa = 3.59GKK355 pKa = 10.95IPMPVDD361 pKa = 3.34SKK363 pKa = 10.75KK364 pKa = 10.91IKK366 pKa = 8.76MQYY369 pKa = 10.41ADD371 pKa = 2.65IYY373 pKa = 9.29YY374 pKa = 8.5YY375 pKa = 10.89RR376 pKa = 11.84KK377 pKa = 9.28MYY379 pKa = 10.8YY380 pKa = 10.06DD381 pKa = 3.48VEE383 pKa = 4.43KK384 pKa = 10.72DD385 pKa = 2.99INGNSRR391 pKa = 11.84YY392 pKa = 9.71VINYY396 pKa = 9.17KK397 pKa = 10.63GIEE400 pKa = 3.97FRR402 pKa = 11.84KK403 pKa = 10.3KK404 pKa = 10.37NLLNEE409 pKa = 4.09LDD411 pKa = 3.68KK412 pKa = 11.6LQLKK416 pKa = 9.21VRR418 pKa = 11.84NCISLVSNSLEE429 pKa = 3.79LQDD432 pKa = 4.61RR433 pKa = 11.84FKK435 pKa = 11.25EE436 pKa = 4.23HH437 pKa = 6.16YY438 pKa = 9.22LKK440 pKa = 10.77CSRR443 pKa = 11.84DD444 pKa = 3.76FYY446 pKa = 11.54FEE448 pKa = 4.94NFDD451 pKa = 4.13KK452 pKa = 11.33NVVLLNFSDD461 pKa = 3.9YY462 pKa = 11.31LYY464 pKa = 10.69INNIDD469 pKa = 3.86NFIEE473 pKa = 4.4KK474 pKa = 9.44YY475 pKa = 10.34AYY477 pKa = 9.63YY478 pKa = 10.77SKK480 pKa = 10.48IYY482 pKa = 10.05RR483 pKa = 11.84NRR485 pKa = 11.84LFRR488 pKa = 11.84QFDD491 pKa = 4.21DD492 pKa = 3.64NFVPLSSYY500 pKa = 10.59VAIDD504 pKa = 4.52PISDD508 pKa = 3.25YY509 pKa = 11.48GRR511 pKa = 11.84FIEE514 pKa = 4.85CRR516 pKa = 11.84VYY518 pKa = 11.14DD519 pKa = 4.7EE520 pKa = 6.31DD521 pKa = 4.34YY522 pKa = 10.99TDD524 pKa = 4.19CFYY527 pKa = 11.38DD528 pKa = 3.48RR529 pKa = 11.84QRR531 pKa = 11.84HH532 pKa = 5.08HH533 pKa = 7.22RR534 pKa = 11.84LHH536 pKa = 7.06LYY538 pKa = 10.5AEE540 pKa = 4.78YY541 pKa = 10.15IKK543 pKa = 10.65HH544 pKa = 6.8PYY546 pKa = 8.26FQEE549 pKa = 4.04IACLCGFFDD558 pKa = 6.09CIQDD562 pKa = 3.97FVDD565 pKa = 3.39KK566 pKa = 11.28LKK568 pKa = 10.97DD569 pKa = 3.27EE570 pKa = 4.45KK571 pKa = 10.66QRR573 pKa = 11.84KK574 pKa = 8.59DD575 pKa = 3.13RR576 pKa = 11.84EE577 pKa = 3.65EE578 pKa = 4.32RR579 pKa = 11.84EE580 pKa = 4.0RR581 pKa = 11.84VKK583 pKa = 11.23ANAYY587 pKa = 10.31SCGLLL592 pKa = 3.59
MM1 pKa = 7.42LTQTLNIRR9 pKa = 11.84NNTHH13 pKa = 5.59YY14 pKa = 10.1FCPSGSRR21 pKa = 11.84LINSVINPSTEE32 pKa = 3.83RR33 pKa = 11.84YY34 pKa = 9.34KK35 pKa = 10.69SLRR38 pKa = 11.84VQGYY42 pKa = 6.98NQRR45 pKa = 11.84LYY47 pKa = 11.38YY48 pKa = 10.29EE49 pKa = 3.92YY50 pKa = 10.05TYY52 pKa = 10.59CKK54 pKa = 9.95SKK56 pKa = 10.67NYY58 pKa = 9.93PVFSYY63 pKa = 10.49TLTYY67 pKa = 10.59NNKK70 pKa = 9.25SLPMFYY76 pKa = 11.08GNAVHH81 pKa = 7.67DD82 pKa = 4.59YY83 pKa = 10.84NDD85 pKa = 2.45IRR87 pKa = 11.84YY88 pKa = 8.08FFRR91 pKa = 11.84EE92 pKa = 4.01SGFDD96 pKa = 3.23KK97 pKa = 10.87HH98 pKa = 6.69LLRR101 pKa = 11.84HH102 pKa = 5.64YY103 pKa = 10.69GSVVKK108 pKa = 9.98YY109 pKa = 8.81FVASEE114 pKa = 4.09LGEE117 pKa = 4.34GGISHH122 pKa = 6.8QDD124 pKa = 3.29SLSHH128 pKa = 5.82NNKK131 pKa = 9.16RR132 pKa = 11.84AQRR135 pKa = 11.84GKK137 pKa = 10.59DD138 pKa = 3.51NNPHH142 pKa = 4.87YY143 pKa = 10.7HH144 pKa = 6.35VIFFVYY150 pKa = 9.94NANDD154 pKa = 2.99TRR156 pKa = 11.84YY157 pKa = 9.71PYY159 pKa = 10.49IRR161 pKa = 11.84INGKK165 pKa = 6.96QFKK168 pKa = 9.89TIVQRR173 pKa = 11.84YY174 pKa = 4.38WQGDD178 pKa = 2.72AWRR181 pKa = 11.84TKK183 pKa = 10.33KK184 pKa = 9.52PQEE187 pKa = 4.04YY188 pKa = 10.19KK189 pKa = 10.84FGIAMNAKK197 pKa = 9.12YY198 pKa = 10.65NLGEE202 pKa = 4.1ITNGSAIRR210 pKa = 11.84YY211 pKa = 6.18NSKK214 pKa = 10.86YY215 pKa = 9.39VVKK218 pKa = 10.65DD219 pKa = 3.16VLQSRR224 pKa = 11.84KK225 pKa = 9.88DD226 pKa = 3.7DD227 pKa = 3.57SCLRR231 pKa = 11.84SCEE234 pKa = 4.36EE235 pKa = 3.8YY236 pKa = 9.75WNSVDD241 pKa = 3.45GEE243 pKa = 4.46YY244 pKa = 10.51FITHH248 pKa = 6.72HH249 pKa = 6.65KK250 pKa = 10.4LFWDD254 pKa = 4.44DD255 pKa = 3.57YY256 pKa = 10.06FDD258 pKa = 5.62RR259 pKa = 11.84YY260 pKa = 11.27VNDD263 pKa = 3.63FASDD267 pKa = 3.42VEE269 pKa = 4.6YY270 pKa = 10.74INGISDD276 pKa = 3.59VFGGFHH282 pKa = 6.62YY283 pKa = 10.5EE284 pKa = 3.94VADD287 pKa = 4.13FSGMSEE293 pKa = 4.02NEE295 pKa = 3.91LYY297 pKa = 10.61DD298 pKa = 3.9AYY300 pKa = 11.42VYY302 pKa = 10.32IYY304 pKa = 10.29GDD306 pKa = 3.17RR307 pKa = 11.84VNFDD311 pKa = 3.45NLLTEE316 pKa = 4.15HH317 pKa = 7.4RR318 pKa = 11.84DD319 pKa = 3.65YY320 pKa = 11.14IIDD323 pKa = 4.37LEE325 pKa = 4.16INRR328 pKa = 11.84YY329 pKa = 8.49KK330 pKa = 10.57NRR332 pKa = 11.84HH333 pKa = 4.14RR334 pKa = 11.84TKK336 pKa = 10.52VKK338 pKa = 9.07ISNGIGLSALDD349 pKa = 3.87KK350 pKa = 10.68MVDD353 pKa = 3.59GKK355 pKa = 10.95IPMPVDD361 pKa = 3.34SKK363 pKa = 10.75KK364 pKa = 10.91IKK366 pKa = 8.76MQYY369 pKa = 10.41ADD371 pKa = 2.65IYY373 pKa = 9.29YY374 pKa = 8.5YY375 pKa = 10.89RR376 pKa = 11.84KK377 pKa = 9.28MYY379 pKa = 10.8YY380 pKa = 10.06DD381 pKa = 3.48VEE383 pKa = 4.43KK384 pKa = 10.72DD385 pKa = 2.99INGNSRR391 pKa = 11.84YY392 pKa = 9.71VINYY396 pKa = 9.17KK397 pKa = 10.63GIEE400 pKa = 3.97FRR402 pKa = 11.84KK403 pKa = 10.3KK404 pKa = 10.37NLLNEE409 pKa = 4.09LDD411 pKa = 3.68KK412 pKa = 11.6LQLKK416 pKa = 9.21VRR418 pKa = 11.84NCISLVSNSLEE429 pKa = 3.79LQDD432 pKa = 4.61RR433 pKa = 11.84FKK435 pKa = 11.25EE436 pKa = 4.23HH437 pKa = 6.16YY438 pKa = 9.22LKK440 pKa = 10.77CSRR443 pKa = 11.84DD444 pKa = 3.76FYY446 pKa = 11.54FEE448 pKa = 4.94NFDD451 pKa = 4.13KK452 pKa = 11.33NVVLLNFSDD461 pKa = 3.9YY462 pKa = 11.31LYY464 pKa = 10.69INNIDD469 pKa = 3.86NFIEE473 pKa = 4.4KK474 pKa = 9.44YY475 pKa = 10.34AYY477 pKa = 9.63YY478 pKa = 10.77SKK480 pKa = 10.48IYY482 pKa = 10.05RR483 pKa = 11.84NRR485 pKa = 11.84LFRR488 pKa = 11.84QFDD491 pKa = 4.21DD492 pKa = 3.64NFVPLSSYY500 pKa = 10.59VAIDD504 pKa = 4.52PISDD508 pKa = 3.25YY509 pKa = 11.48GRR511 pKa = 11.84FIEE514 pKa = 4.85CRR516 pKa = 11.84VYY518 pKa = 11.14DD519 pKa = 4.7EE520 pKa = 6.31DD521 pKa = 4.34YY522 pKa = 10.99TDD524 pKa = 4.19CFYY527 pKa = 11.38DD528 pKa = 3.48RR529 pKa = 11.84QRR531 pKa = 11.84HH532 pKa = 5.08HH533 pKa = 7.22RR534 pKa = 11.84LHH536 pKa = 7.06LYY538 pKa = 10.5AEE540 pKa = 4.78YY541 pKa = 10.15IKK543 pKa = 10.65HH544 pKa = 6.8PYY546 pKa = 8.26FQEE549 pKa = 4.04IACLCGFFDD558 pKa = 6.09CIQDD562 pKa = 3.97FVDD565 pKa = 3.39KK566 pKa = 11.28LKK568 pKa = 10.97DD569 pKa = 3.27EE570 pKa = 4.45KK571 pKa = 10.66QRR573 pKa = 11.84KK574 pKa = 8.59DD575 pKa = 3.13RR576 pKa = 11.84EE577 pKa = 3.65EE578 pKa = 4.32RR579 pKa = 11.84EE580 pKa = 4.0RR581 pKa = 11.84VKK583 pKa = 11.23ANAYY587 pKa = 10.31SCGLLL592 pKa = 3.59
Molecular weight: 71.11 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1671 |
107 |
592 |
417.8 |
47.8 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.925 ± 1.683 | 0.898 ± 0.495 |
7.54 ± 0.332 | 4.608 ± 0.388 |
4.967 ± 0.419 | 6.344 ± 0.783 |
1.855 ± 0.568 | 5.745 ± 0.425 |
6.523 ± 0.544 | 7.241 ± 0.171 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.334 ± 0.465 | 7.959 ± 0.614 |
2.454 ± 0.317 | 4.189 ± 0.788 |
4.668 ± 1.039 | 7.899 ± 0.724 |
4.907 ± 1.12 | 6.104 ± 0.473 |
0.958 ± 0.151 | 6.882 ± 1.5 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
