
Tortoise microvirus 96
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 6.89
Get precalculated fractions of proteins
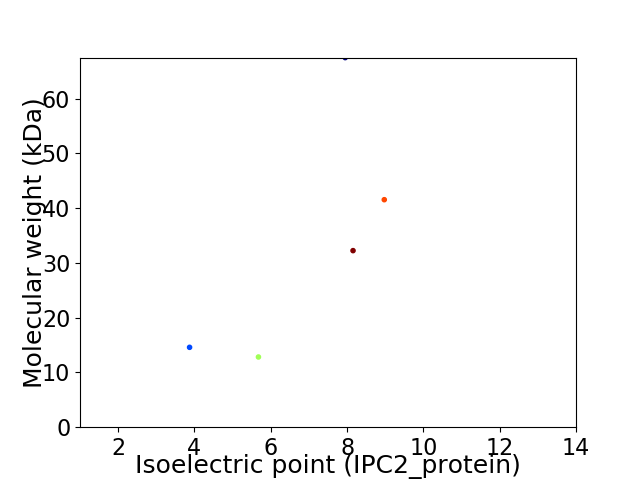
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4P8W7P8|A0A4P8W7P8_9VIRU DNA pilot protein OS=Tortoise microvirus 96 OX=2583205 PE=4 SV=1
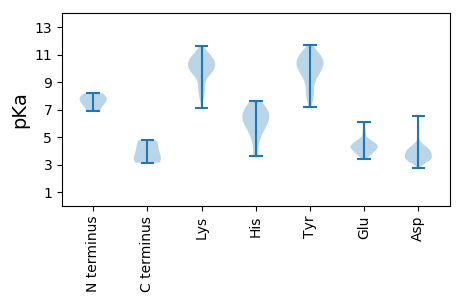
MM1 pKa = 8.18PINICPTARR10 pKa = 11.84EE11 pKa = 3.96PQVLDD16 pKa = 3.98FEE18 pKa = 5.3DD19 pKa = 4.39SVVEE23 pKa = 3.9PDD25 pKa = 3.14EE26 pKa = 4.69AMSVTDD32 pKa = 4.08IIRR35 pKa = 11.84RR36 pKa = 11.84HH37 pKa = 6.14LSGQPLSVPGDD48 pKa = 3.54YY49 pKa = 11.05DD50 pKa = 3.59GTDD53 pKa = 3.15VGDD56 pKa = 4.14NEE58 pKa = 5.93LDD60 pKa = 4.04DD61 pKa = 5.83SDD63 pKa = 6.53DD64 pKa = 3.79EE65 pKa = 4.35MFDD68 pKa = 5.08AISQRR73 pKa = 11.84IDD75 pKa = 3.43AMDD78 pKa = 3.57LTEE81 pKa = 4.71IDD83 pKa = 3.96EE84 pKa = 4.33MRR86 pKa = 11.84SMLNNRR92 pKa = 11.84AASIRR97 pKa = 11.84AQLKK101 pKa = 8.49AQEE104 pKa = 4.27DD105 pKa = 3.97AALKK109 pKa = 9.62QQIKK113 pKa = 11.2DD114 pKa = 3.61EE115 pKa = 4.25MEE117 pKa = 4.64KK118 pKa = 10.78DD119 pKa = 3.87SQSDD123 pKa = 3.59PLDD126 pKa = 3.84NSHH129 pKa = 7.38PAA131 pKa = 3.14
MM1 pKa = 8.18PINICPTARR10 pKa = 11.84EE11 pKa = 3.96PQVLDD16 pKa = 3.98FEE18 pKa = 5.3DD19 pKa = 4.39SVVEE23 pKa = 3.9PDD25 pKa = 3.14EE26 pKa = 4.69AMSVTDD32 pKa = 4.08IIRR35 pKa = 11.84RR36 pKa = 11.84HH37 pKa = 6.14LSGQPLSVPGDD48 pKa = 3.54YY49 pKa = 11.05DD50 pKa = 3.59GTDD53 pKa = 3.15VGDD56 pKa = 4.14NEE58 pKa = 5.93LDD60 pKa = 4.04DD61 pKa = 5.83SDD63 pKa = 6.53DD64 pKa = 3.79EE65 pKa = 4.35MFDD68 pKa = 5.08AISQRR73 pKa = 11.84IDD75 pKa = 3.43AMDD78 pKa = 3.57LTEE81 pKa = 4.71IDD83 pKa = 3.96EE84 pKa = 4.33MRR86 pKa = 11.84SMLNNRR92 pKa = 11.84AASIRR97 pKa = 11.84AQLKK101 pKa = 8.49AQEE104 pKa = 4.27DD105 pKa = 3.97AALKK109 pKa = 9.62QQIKK113 pKa = 11.2DD114 pKa = 3.61EE115 pKa = 4.25MEE117 pKa = 4.64KK118 pKa = 10.78DD119 pKa = 3.87SQSDD123 pKa = 3.59PLDD126 pKa = 3.84NSHH129 pKa = 7.38PAA131 pKa = 3.14
Molecular weight: 14.59 kDa
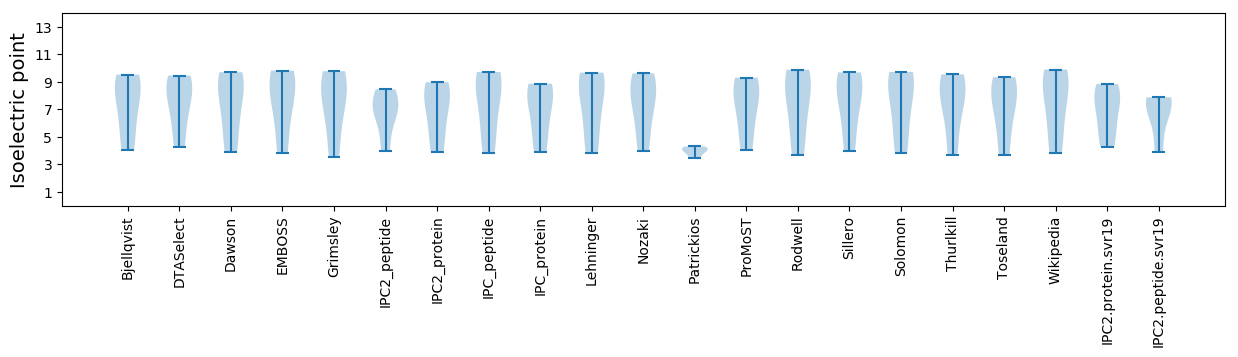
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4V1FW21|A0A4V1FW21_9VIRU Uncharacterized protein OS=Tortoise microvirus 96 OX=2583205 PE=4 SV=1
MM1 pKa = 7.85LINNMLNFKK10 pKa = 8.04TFKK13 pKa = 9.95LWLIVVVVAAAAVAAFVVTAFRR35 pKa = 11.84VEE37 pKa = 3.89ASVCNKK43 pKa = 9.55QLIIMNCLSPIRR55 pKa = 11.84IKK57 pKa = 10.87NPKK60 pKa = 7.11PTYY63 pKa = 8.33YY64 pKa = 10.39SPRR67 pKa = 11.84NFIDD71 pKa = 4.14VPCGKK76 pKa = 9.81CVHH79 pKa = 6.22CLKK82 pKa = 10.58NKK84 pKa = 9.67RR85 pKa = 11.84RR86 pKa = 11.84EE87 pKa = 3.91LSFRR91 pKa = 11.84IYY93 pKa = 11.1NEE95 pKa = 3.66LRR97 pKa = 11.84DD98 pKa = 3.83AEE100 pKa = 4.25SAYY103 pKa = 10.49FVTLTYY109 pKa = 7.79DD110 pKa = 3.5TPNLQFNLHH119 pKa = 6.41SGLPILVKK127 pKa = 10.3RR128 pKa = 11.84DD129 pKa = 3.21VQLFLKK135 pKa = 10.43RR136 pKa = 11.84LRR138 pKa = 11.84KK139 pKa = 9.72DD140 pKa = 3.42LSSKK144 pKa = 10.95GLDD147 pKa = 3.42HH148 pKa = 7.56KK149 pKa = 10.66NLKK152 pKa = 8.86YY153 pKa = 10.72FCVSEE158 pKa = 4.43FGDD161 pKa = 3.5EE162 pKa = 4.23FMRR165 pKa = 11.84PHH167 pKa = 6.69YY168 pKa = 10.85HH169 pKa = 6.54MILFNLPIQDD179 pKa = 3.88IPVTTAKK186 pKa = 9.9RR187 pKa = 11.84CYY189 pKa = 9.88YY190 pKa = 9.83PVIKK194 pKa = 10.36QYY196 pKa = 11.31LDD198 pKa = 4.5SIWQLGFTDD207 pKa = 4.11VGCVTLASCNYY218 pKa = 8.02CAKK221 pKa = 9.92YY222 pKa = 9.49VCKK225 pKa = 10.53GLFNLDD231 pKa = 3.19EE232 pKa = 4.73KK233 pKa = 10.96PWVNEE238 pKa = 3.39YY239 pKa = 7.2WTLRR243 pKa = 11.84SQGIGLNFVDD253 pKa = 4.43KK254 pKa = 11.02YY255 pKa = 11.27GNRR258 pKa = 11.84FLKK261 pKa = 10.5SGKK264 pKa = 8.17TFTTWHH270 pKa = 6.2SFPIKK275 pKa = 10.26LPRR278 pKa = 11.84YY279 pKa = 8.03YY280 pKa = 10.06INKK283 pKa = 8.96IFYY286 pKa = 11.01NNDD289 pKa = 2.82SDD291 pKa = 5.6NFHH294 pKa = 4.95QHH296 pKa = 5.82CRR298 pKa = 11.84LSSKK302 pKa = 9.79AWNYY306 pKa = 10.98NNTAGNDD313 pKa = 3.3AYY315 pKa = 11.37SYY317 pKa = 10.7FLRR320 pKa = 11.84ASNRR324 pKa = 11.84QFEE327 pKa = 4.22NAAILFGQYY336 pKa = 7.44QHH338 pKa = 6.44ATVNNVLRR346 pKa = 11.84ASGGFRR352 pKa = 11.84ITNVRR357 pKa = 3.32
MM1 pKa = 7.85LINNMLNFKK10 pKa = 8.04TFKK13 pKa = 9.95LWLIVVVVAAAAVAAFVVTAFRR35 pKa = 11.84VEE37 pKa = 3.89ASVCNKK43 pKa = 9.55QLIIMNCLSPIRR55 pKa = 11.84IKK57 pKa = 10.87NPKK60 pKa = 7.11PTYY63 pKa = 8.33YY64 pKa = 10.39SPRR67 pKa = 11.84NFIDD71 pKa = 4.14VPCGKK76 pKa = 9.81CVHH79 pKa = 6.22CLKK82 pKa = 10.58NKK84 pKa = 9.67RR85 pKa = 11.84RR86 pKa = 11.84EE87 pKa = 3.91LSFRR91 pKa = 11.84IYY93 pKa = 11.1NEE95 pKa = 3.66LRR97 pKa = 11.84DD98 pKa = 3.83AEE100 pKa = 4.25SAYY103 pKa = 10.49FVTLTYY109 pKa = 7.79DD110 pKa = 3.5TPNLQFNLHH119 pKa = 6.41SGLPILVKK127 pKa = 10.3RR128 pKa = 11.84DD129 pKa = 3.21VQLFLKK135 pKa = 10.43RR136 pKa = 11.84LRR138 pKa = 11.84KK139 pKa = 9.72DD140 pKa = 3.42LSSKK144 pKa = 10.95GLDD147 pKa = 3.42HH148 pKa = 7.56KK149 pKa = 10.66NLKK152 pKa = 8.86YY153 pKa = 10.72FCVSEE158 pKa = 4.43FGDD161 pKa = 3.5EE162 pKa = 4.23FMRR165 pKa = 11.84PHH167 pKa = 6.69YY168 pKa = 10.85HH169 pKa = 6.54MILFNLPIQDD179 pKa = 3.88IPVTTAKK186 pKa = 9.9RR187 pKa = 11.84CYY189 pKa = 9.88YY190 pKa = 9.83PVIKK194 pKa = 10.36QYY196 pKa = 11.31LDD198 pKa = 4.5SIWQLGFTDD207 pKa = 4.11VGCVTLASCNYY218 pKa = 8.02CAKK221 pKa = 9.92YY222 pKa = 9.49VCKK225 pKa = 10.53GLFNLDD231 pKa = 3.19EE232 pKa = 4.73KK233 pKa = 10.96PWVNEE238 pKa = 3.39YY239 pKa = 7.2WTLRR243 pKa = 11.84SQGIGLNFVDD253 pKa = 4.43KK254 pKa = 11.02YY255 pKa = 11.27GNRR258 pKa = 11.84FLKK261 pKa = 10.5SGKK264 pKa = 8.17TFTTWHH270 pKa = 6.2SFPIKK275 pKa = 10.26LPRR278 pKa = 11.84YY279 pKa = 8.03YY280 pKa = 10.06INKK283 pKa = 8.96IFYY286 pKa = 11.01NNDD289 pKa = 2.82SDD291 pKa = 5.6NFHH294 pKa = 4.95QHH296 pKa = 5.82CRR298 pKa = 11.84LSSKK302 pKa = 9.79AWNYY306 pKa = 10.98NNTAGNDD313 pKa = 3.3AYY315 pKa = 11.37SYY317 pKa = 10.7FLRR320 pKa = 11.84ASNRR324 pKa = 11.84QFEE327 pKa = 4.22NAAILFGQYY336 pKa = 7.44QHH338 pKa = 6.44ATVNNVLRR346 pKa = 11.84ASGGFRR352 pKa = 11.84ITNVRR357 pKa = 3.32
Molecular weight: 41.56 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1481 |
112 |
593 |
296.2 |
33.75 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.617 ± 1.169 | 1.215 ± 0.584 |
5.807 ± 1.718 | 4.186 ± 0.724 |
5.739 ± 0.929 | 5.874 ± 0.873 |
2.026 ± 0.569 | 5.672 ± 0.503 |
4.727 ± 0.73 | 8.778 ± 0.755 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.566 ± 0.576 | 6.955 ± 1.466 |
4.929 ± 0.915 | 5.199 ± 1.134 |
5.537 ± 0.252 | 7.427 ± 0.88 |
4.862 ± 0.58 | 5.604 ± 0.916 |
1.148 ± 0.3 | 5.132 ± 0.688 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
