
Thrips-associated genomovirus 4
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Genomoviridae; Gemykolovirus; Gemykolovirus echia1
Average proteome isoelectric point is 6.95
Get precalculated fractions of proteins
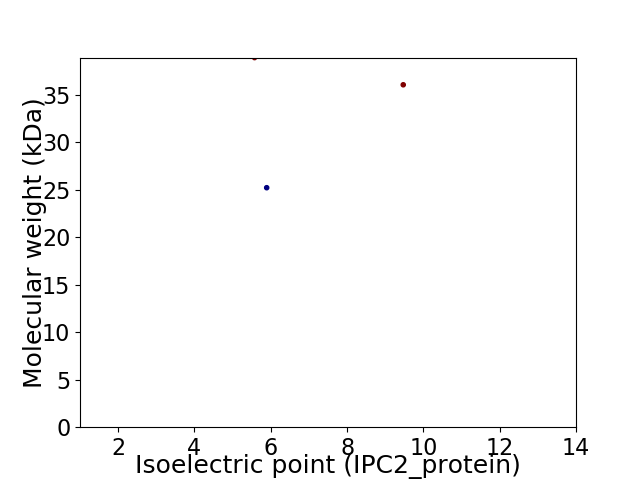
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1P8YT69|A0A1P8YT69_9VIRU Replication associated protein OS=Thrips-associated genomovirus 4 OX=1941239 PE=4 SV=1
MM1 pKa = 7.1VRR3 pKa = 11.84RR4 pKa = 11.84FKK6 pKa = 10.58IDD8 pKa = 3.41DD9 pKa = 3.66EE10 pKa = 4.63QFFMLTYY17 pKa = 7.77PTTPDD22 pKa = 4.27DD23 pKa = 4.78FPIDD27 pKa = 3.99DD28 pKa = 4.82LVGILDD34 pKa = 4.16GLGCNYY40 pKa = 10.49RR41 pKa = 11.84IGRR44 pKa = 11.84EE45 pKa = 3.77LHH47 pKa = 6.79EE48 pKa = 5.17DD49 pKa = 3.94GKK51 pKa = 9.81PHH53 pKa = 5.72FHH55 pKa = 7.36AMVVFDD61 pKa = 4.8NPYY64 pKa = 9.56SDD66 pKa = 3.78RR67 pKa = 11.84DD68 pKa = 3.31GRR70 pKa = 11.84TTFRR74 pKa = 11.84VGGRR78 pKa = 11.84APNIRR83 pKa = 11.84MRR85 pKa = 11.84RR86 pKa = 11.84TKK88 pKa = 10.29PEE90 pKa = 4.07RR91 pKa = 11.84GWDD94 pKa = 3.64YY95 pKa = 11.11VGKK98 pKa = 9.63HH99 pKa = 5.96AGSKK103 pKa = 9.51DD104 pKa = 2.89GHH106 pKa = 6.41YY107 pKa = 10.36IVAEE111 pKa = 4.21KK112 pKa = 10.59GDD114 pKa = 3.86RR115 pKa = 11.84PGGDD119 pKa = 3.21GDD121 pKa = 4.68SNDD124 pKa = 3.77RR125 pKa = 11.84STNDD129 pKa = 2.62VWHH132 pKa = 6.8EE133 pKa = 4.01IILARR138 pKa = 11.84TRR140 pKa = 11.84EE141 pKa = 4.06EE142 pKa = 4.21FFDD145 pKa = 3.96LASRR149 pKa = 11.84LAPRR153 pKa = 11.84QLACSFTQLTSYY165 pKa = 11.32ADD167 pKa = 2.93WKK169 pKa = 9.97YY170 pKa = 10.64RR171 pKa = 11.84VVEE174 pKa = 4.19TPYY177 pKa = 11.01ASPEE181 pKa = 4.21GEE183 pKa = 3.73FEE185 pKa = 4.72VPIEE189 pKa = 3.8LQQWVNANLRR199 pKa = 11.84RR200 pKa = 11.84EE201 pKa = 3.98HH202 pKa = 6.65HH203 pKa = 6.12GRR205 pKa = 11.84PNGLVLFGATRR216 pKa = 11.84LGKK219 pKa = 7.65TVWARR224 pKa = 11.84SLGQHH229 pKa = 6.23YY230 pKa = 10.39YY231 pKa = 10.66CAGLWDD237 pKa = 4.47LSRR240 pKa = 11.84FDD242 pKa = 4.29EE243 pKa = 4.4SVEE246 pKa = 3.8YY247 pKa = 10.63AIFDD251 pKa = 4.02DD252 pKa = 4.08MVGGLRR258 pKa = 11.84AGYY261 pKa = 10.01FNYY264 pKa = 10.23KK265 pKa = 10.05DD266 pKa = 3.2WLGGQFEE273 pKa = 4.66FTVQDD278 pKa = 3.37KK279 pKa = 10.87YY280 pKa = 11.3KK281 pKa = 10.46HH282 pKa = 5.91KK283 pKa = 10.9KK284 pKa = 7.53QIKK287 pKa = 7.04WGKK290 pKa = 7.32PAIYY294 pKa = 9.74ICNQDD299 pKa = 3.51PRR301 pKa = 11.84TDD303 pKa = 3.21ITPMGKK309 pKa = 9.02NAIEE313 pKa = 3.99WDD315 pKa = 3.47WMEE318 pKa = 4.25EE319 pKa = 3.61NCVFYY324 pKa = 10.78EE325 pKa = 4.12CTEE328 pKa = 4.32TIFRR332 pKa = 11.84ASTT335 pKa = 3.11
MM1 pKa = 7.1VRR3 pKa = 11.84RR4 pKa = 11.84FKK6 pKa = 10.58IDD8 pKa = 3.41DD9 pKa = 3.66EE10 pKa = 4.63QFFMLTYY17 pKa = 7.77PTTPDD22 pKa = 4.27DD23 pKa = 4.78FPIDD27 pKa = 3.99DD28 pKa = 4.82LVGILDD34 pKa = 4.16GLGCNYY40 pKa = 10.49RR41 pKa = 11.84IGRR44 pKa = 11.84EE45 pKa = 3.77LHH47 pKa = 6.79EE48 pKa = 5.17DD49 pKa = 3.94GKK51 pKa = 9.81PHH53 pKa = 5.72FHH55 pKa = 7.36AMVVFDD61 pKa = 4.8NPYY64 pKa = 9.56SDD66 pKa = 3.78RR67 pKa = 11.84DD68 pKa = 3.31GRR70 pKa = 11.84TTFRR74 pKa = 11.84VGGRR78 pKa = 11.84APNIRR83 pKa = 11.84MRR85 pKa = 11.84RR86 pKa = 11.84TKK88 pKa = 10.29PEE90 pKa = 4.07RR91 pKa = 11.84GWDD94 pKa = 3.64YY95 pKa = 11.11VGKK98 pKa = 9.63HH99 pKa = 5.96AGSKK103 pKa = 9.51DD104 pKa = 2.89GHH106 pKa = 6.41YY107 pKa = 10.36IVAEE111 pKa = 4.21KK112 pKa = 10.59GDD114 pKa = 3.86RR115 pKa = 11.84PGGDD119 pKa = 3.21GDD121 pKa = 4.68SNDD124 pKa = 3.77RR125 pKa = 11.84STNDD129 pKa = 2.62VWHH132 pKa = 6.8EE133 pKa = 4.01IILARR138 pKa = 11.84TRR140 pKa = 11.84EE141 pKa = 4.06EE142 pKa = 4.21FFDD145 pKa = 3.96LASRR149 pKa = 11.84LAPRR153 pKa = 11.84QLACSFTQLTSYY165 pKa = 11.32ADD167 pKa = 2.93WKK169 pKa = 9.97YY170 pKa = 10.64RR171 pKa = 11.84VVEE174 pKa = 4.19TPYY177 pKa = 11.01ASPEE181 pKa = 4.21GEE183 pKa = 3.73FEE185 pKa = 4.72VPIEE189 pKa = 3.8LQQWVNANLRR199 pKa = 11.84RR200 pKa = 11.84EE201 pKa = 3.98HH202 pKa = 6.65HH203 pKa = 6.12GRR205 pKa = 11.84PNGLVLFGATRR216 pKa = 11.84LGKK219 pKa = 7.65TVWARR224 pKa = 11.84SLGQHH229 pKa = 6.23YY230 pKa = 10.39YY231 pKa = 10.66CAGLWDD237 pKa = 4.47LSRR240 pKa = 11.84FDD242 pKa = 4.29EE243 pKa = 4.4SVEE246 pKa = 3.8YY247 pKa = 10.63AIFDD251 pKa = 4.02DD252 pKa = 4.08MVGGLRR258 pKa = 11.84AGYY261 pKa = 10.01FNYY264 pKa = 10.23KK265 pKa = 10.05DD266 pKa = 3.2WLGGQFEE273 pKa = 4.66FTVQDD278 pKa = 3.37KK279 pKa = 10.87YY280 pKa = 11.3KK281 pKa = 10.46HH282 pKa = 5.91KK283 pKa = 10.9KK284 pKa = 7.53QIKK287 pKa = 7.04WGKK290 pKa = 7.32PAIYY294 pKa = 9.74ICNQDD299 pKa = 3.51PRR301 pKa = 11.84TDD303 pKa = 3.21ITPMGKK309 pKa = 9.02NAIEE313 pKa = 3.99WDD315 pKa = 3.47WMEE318 pKa = 4.25EE319 pKa = 3.61NCVFYY324 pKa = 10.78EE325 pKa = 4.12CTEE328 pKa = 4.32TIFRR332 pKa = 11.84ASTT335 pKa = 3.11
Molecular weight: 38.89 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1P8YT69|A0A1P8YT69_9VIRU Replication associated protein OS=Thrips-associated genomovirus 4 OX=1941239 PE=4 SV=1
MM1 pKa = 7.54AYY3 pKa = 9.97SRR5 pKa = 11.84RR6 pKa = 11.84RR7 pKa = 11.84YY8 pKa = 9.66GSSRR12 pKa = 11.84AKK14 pKa = 9.64KK15 pKa = 4.61TTKK18 pKa = 9.96RR19 pKa = 11.84SGRR22 pKa = 11.84RR23 pKa = 11.84SRR25 pKa = 11.84YY26 pKa = 4.92TAKK29 pKa = 9.55TRR31 pKa = 11.84RR32 pKa = 11.84YY33 pKa = 9.34KK34 pKa = 10.73KK35 pKa = 8.69KK36 pKa = 8.14TYY38 pKa = 10.14RR39 pKa = 11.84KK40 pKa = 10.08ASMKK44 pKa = 10.18TILNKK49 pKa = 9.31TSRR52 pKa = 11.84KK53 pKa = 8.91KK54 pKa = 10.58RR55 pKa = 11.84NGMLTFANSTATGALATVSQAPLTIAGSPSGDD87 pKa = 3.24LRR89 pKa = 11.84GLIHH93 pKa = 7.06FRR95 pKa = 11.84PTSMDD100 pKa = 3.62LDD102 pKa = 3.71DD103 pKa = 4.91TVTNPNSIISTPSRR117 pKa = 11.84TASVCYY123 pKa = 9.54MKK125 pKa = 10.99GFAEE129 pKa = 4.13NVRR132 pKa = 11.84VEE134 pKa = 4.33TSTGNPWFHH143 pKa = 6.7RR144 pKa = 11.84RR145 pKa = 11.84ICITVKK151 pKa = 10.64HH152 pKa = 5.94PLFYY156 pKa = 11.08ALDD159 pKa = 3.77SRR161 pKa = 11.84DD162 pKa = 3.64TNGTDD167 pKa = 2.57RR168 pKa = 11.84LYY170 pKa = 10.88AARR173 pKa = 11.84GAIEE177 pKa = 4.47TSNGWQRR184 pKa = 11.84LASNVMTDD192 pKa = 3.63TLSNTYY198 pKa = 10.31GGWLSVMFKK207 pKa = 10.61GVQGVDD213 pKa = 2.49WDD215 pKa = 4.43DD216 pKa = 5.63FITAPIDD223 pKa = 3.49TNRR226 pKa = 11.84VDD228 pKa = 5.11LKK230 pKa = 11.0FDD232 pKa = 3.4KK233 pKa = 10.32TWVYY237 pKa = 10.84RR238 pKa = 11.84SGNEE242 pKa = 3.52RR243 pKa = 11.84GVLKK247 pKa = 9.03EE248 pKa = 4.08TKK250 pKa = 9.3LWHH253 pKa = 6.43PMNKK257 pKa = 9.35NLYY260 pKa = 9.28YY261 pKa = 10.79DD262 pKa = 4.61DD263 pKa = 5.72DD264 pKa = 4.52EE265 pKa = 6.6SGATEE270 pKa = 4.03EE271 pKa = 4.18TRR273 pKa = 11.84TWSVLDD279 pKa = 3.42KK280 pKa = 11.07RR281 pKa = 11.84GMGDD285 pKa = 2.98YY286 pKa = 10.63HH287 pKa = 8.66IFDD290 pKa = 5.17LFSQGSSGATTDD302 pKa = 3.61LLKK305 pKa = 10.47IRR307 pKa = 11.84FTSTMYY313 pKa = 9.45WHH315 pKa = 7.08EE316 pKa = 4.17KK317 pKa = 9.5
MM1 pKa = 7.54AYY3 pKa = 9.97SRR5 pKa = 11.84RR6 pKa = 11.84RR7 pKa = 11.84YY8 pKa = 9.66GSSRR12 pKa = 11.84AKK14 pKa = 9.64KK15 pKa = 4.61TTKK18 pKa = 9.96RR19 pKa = 11.84SGRR22 pKa = 11.84RR23 pKa = 11.84SRR25 pKa = 11.84YY26 pKa = 4.92TAKK29 pKa = 9.55TRR31 pKa = 11.84RR32 pKa = 11.84YY33 pKa = 9.34KK34 pKa = 10.73KK35 pKa = 8.69KK36 pKa = 8.14TYY38 pKa = 10.14RR39 pKa = 11.84KK40 pKa = 10.08ASMKK44 pKa = 10.18TILNKK49 pKa = 9.31TSRR52 pKa = 11.84KK53 pKa = 8.91KK54 pKa = 10.58RR55 pKa = 11.84NGMLTFANSTATGALATVSQAPLTIAGSPSGDD87 pKa = 3.24LRR89 pKa = 11.84GLIHH93 pKa = 7.06FRR95 pKa = 11.84PTSMDD100 pKa = 3.62LDD102 pKa = 3.71DD103 pKa = 4.91TVTNPNSIISTPSRR117 pKa = 11.84TASVCYY123 pKa = 9.54MKK125 pKa = 10.99GFAEE129 pKa = 4.13NVRR132 pKa = 11.84VEE134 pKa = 4.33TSTGNPWFHH143 pKa = 6.7RR144 pKa = 11.84RR145 pKa = 11.84ICITVKK151 pKa = 10.64HH152 pKa = 5.94PLFYY156 pKa = 11.08ALDD159 pKa = 3.77SRR161 pKa = 11.84DD162 pKa = 3.64TNGTDD167 pKa = 2.57RR168 pKa = 11.84LYY170 pKa = 10.88AARR173 pKa = 11.84GAIEE177 pKa = 4.47TSNGWQRR184 pKa = 11.84LASNVMTDD192 pKa = 3.63TLSNTYY198 pKa = 10.31GGWLSVMFKK207 pKa = 10.61GVQGVDD213 pKa = 2.49WDD215 pKa = 4.43DD216 pKa = 5.63FITAPIDD223 pKa = 3.49TNRR226 pKa = 11.84VDD228 pKa = 5.11LKK230 pKa = 11.0FDD232 pKa = 3.4KK233 pKa = 10.32TWVYY237 pKa = 10.84RR238 pKa = 11.84SGNEE242 pKa = 3.52RR243 pKa = 11.84GVLKK247 pKa = 9.03EE248 pKa = 4.08TKK250 pKa = 9.3LWHH253 pKa = 6.43PMNKK257 pKa = 9.35NLYY260 pKa = 9.28YY261 pKa = 10.79DD262 pKa = 4.61DD263 pKa = 5.72DD264 pKa = 4.52EE265 pKa = 6.6SGATEE270 pKa = 4.03EE271 pKa = 4.18TRR273 pKa = 11.84TWSVLDD279 pKa = 3.42KK280 pKa = 11.07RR281 pKa = 11.84GMGDD285 pKa = 2.98YY286 pKa = 10.63HH287 pKa = 8.66IFDD290 pKa = 5.17LFSQGSSGATTDD302 pKa = 3.61LLKK305 pKa = 10.47IRR307 pKa = 11.84FTSTMYY313 pKa = 9.45WHH315 pKa = 7.08EE316 pKa = 4.17KK317 pKa = 9.5
Molecular weight: 36.03 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
871 |
219 |
335 |
290.3 |
33.37 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.855 ± 0.194 | 1.148 ± 0.3 |
8.037 ± 0.598 | 5.166 ± 0.962 |
4.707 ± 0.532 | 8.496 ± 0.539 |
2.755 ± 0.396 | 4.248 ± 0.249 |
5.281 ± 0.911 | 6.315 ± 0.163 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.411 ± 0.315 | 3.904 ± 0.357 |
4.478 ± 0.792 | 2.296 ± 0.431 |
9.3 ± 0.641 | 5.741 ± 1.412 |
7.692 ± 1.656 | 5.166 ± 0.342 |
2.526 ± 0.283 | 4.478 ± 0.164 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
