
Tessaracoccus lapidicaptus
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Propionibacteriales; Propionibacteriaceae; Tessaracoccus
Average proteome isoelectric point is 6.01
Get precalculated fractions of proteins
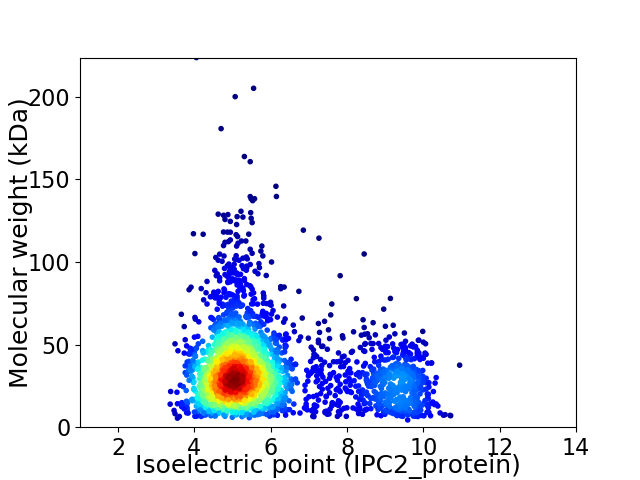
Virtual 2D-PAGE plot for 2736 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1C0ARM4|A0A1C0ARM4_9ACTN HTH-type transcriptional regulator LutR OS=Tessaracoccus lapidicaptus OX=1427523 GN=lutR_4 PE=4 SV=1
MM1 pKa = 7.27ATFNFRR7 pKa = 11.84KK8 pKa = 10.26AGTALAAVTALSLTLTACGGEE29 pKa = 4.28SEE31 pKa = 4.4PAATNGGDD39 pKa = 3.64GEE41 pKa = 4.65AVDD44 pKa = 4.31TSAASGQIDD53 pKa = 3.7YY54 pKa = 9.89WLWDD58 pKa = 3.8NNQKK62 pKa = 9.77PAYY65 pKa = 6.68EE66 pKa = 3.74QCAADD71 pKa = 3.95FQEE74 pKa = 4.31EE75 pKa = 4.54TGVSVTITQYY85 pKa = 11.61AWDD88 pKa = 5.31DD89 pKa = 3.56YY90 pKa = 11.41WSGITNGFVAGTAPDD105 pKa = 3.41VFTNHH110 pKa = 6.82LSRR113 pKa = 11.84YY114 pKa = 8.95PEE116 pKa = 4.31FVTQGQLMALDD127 pKa = 4.07ATLEE131 pKa = 4.16ADD133 pKa = 3.64GVDD136 pKa = 2.95VDD138 pKa = 5.41GYY140 pKa = 10.91QEE142 pKa = 4.08GLADD146 pKa = 4.03LWVGQDD152 pKa = 3.31GLRR155 pKa = 11.84YY156 pKa = 9.62GLPKK160 pKa = 10.94DD161 pKa = 3.74FDD163 pKa = 4.16TVAIFYY169 pKa = 10.44NKK171 pKa = 10.44ALIEE175 pKa = 4.02EE176 pKa = 4.59AGVDD180 pKa = 3.93EE181 pKa = 5.53ADD183 pKa = 4.3LAALEE188 pKa = 4.45WNPEE192 pKa = 3.9DD193 pKa = 3.68GGTYY197 pKa = 10.59EE198 pKa = 5.14EE199 pKa = 5.09MIAHH203 pKa = 6.3LTIDD207 pKa = 3.76NNGVRR212 pKa = 11.84GDD214 pKa = 3.62EE215 pKa = 4.19PGFDD219 pKa = 3.36KK220 pKa = 11.15TKK222 pKa = 9.65VAVYY226 pKa = 10.65GLGLDD231 pKa = 3.96GGSGGGNGQTQWSMYY246 pKa = 10.39SGTTGWTVTDD256 pKa = 3.85KK257 pKa = 11.44NPWGTKK263 pKa = 8.62YY264 pKa = 10.87NYY266 pKa = 10.04DD267 pKa = 3.35AAEE270 pKa = 4.31FKK272 pKa = 10.43DD273 pKa = 4.06TISWMASLAEE283 pKa = 4.25KK284 pKa = 10.67GYY286 pKa = 8.86MPTVEE291 pKa = 4.74AVTGQSSGDD300 pKa = 3.03IFGAGKK306 pKa = 10.33YY307 pKa = 11.05AMITNGSWMINQMFGYY323 pKa = 8.87TGIEE327 pKa = 3.98TGLAPTPVGPSGEE340 pKa = 4.08RR341 pKa = 11.84ASMYY345 pKa = 10.68NGLADD350 pKa = 4.14SVWAGSDD357 pKa = 3.04NKK359 pKa = 10.48AAAVKK364 pKa = 8.48WVEE367 pKa = 4.08YY368 pKa = 10.1LGSAACQDD376 pKa = 3.52IVGEE380 pKa = 4.13AGVVFPAIPSATEE393 pKa = 3.39KK394 pKa = 10.79AQAAFADD401 pKa = 3.92RR402 pKa = 11.84GIDD405 pKa = 3.46VEE407 pKa = 4.28PFLVHH412 pKa = 6.36VNEE415 pKa = 4.25GTTFLFPITDD425 pKa = 3.58YY426 pKa = 11.42ASQINGIMQPAVDD439 pKa = 4.79AVFTGQADD447 pKa = 3.85VDD449 pKa = 4.26SLDD452 pKa = 3.74AANEE456 pKa = 3.97QVNALFNGG464 pKa = 3.79
MM1 pKa = 7.27ATFNFRR7 pKa = 11.84KK8 pKa = 10.26AGTALAAVTALSLTLTACGGEE29 pKa = 4.28SEE31 pKa = 4.4PAATNGGDD39 pKa = 3.64GEE41 pKa = 4.65AVDD44 pKa = 4.31TSAASGQIDD53 pKa = 3.7YY54 pKa = 9.89WLWDD58 pKa = 3.8NNQKK62 pKa = 9.77PAYY65 pKa = 6.68EE66 pKa = 3.74QCAADD71 pKa = 3.95FQEE74 pKa = 4.31EE75 pKa = 4.54TGVSVTITQYY85 pKa = 11.61AWDD88 pKa = 5.31DD89 pKa = 3.56YY90 pKa = 11.41WSGITNGFVAGTAPDD105 pKa = 3.41VFTNHH110 pKa = 6.82LSRR113 pKa = 11.84YY114 pKa = 8.95PEE116 pKa = 4.31FVTQGQLMALDD127 pKa = 4.07ATLEE131 pKa = 4.16ADD133 pKa = 3.64GVDD136 pKa = 2.95VDD138 pKa = 5.41GYY140 pKa = 10.91QEE142 pKa = 4.08GLADD146 pKa = 4.03LWVGQDD152 pKa = 3.31GLRR155 pKa = 11.84YY156 pKa = 9.62GLPKK160 pKa = 10.94DD161 pKa = 3.74FDD163 pKa = 4.16TVAIFYY169 pKa = 10.44NKK171 pKa = 10.44ALIEE175 pKa = 4.02EE176 pKa = 4.59AGVDD180 pKa = 3.93EE181 pKa = 5.53ADD183 pKa = 4.3LAALEE188 pKa = 4.45WNPEE192 pKa = 3.9DD193 pKa = 3.68GGTYY197 pKa = 10.59EE198 pKa = 5.14EE199 pKa = 5.09MIAHH203 pKa = 6.3LTIDD207 pKa = 3.76NNGVRR212 pKa = 11.84GDD214 pKa = 3.62EE215 pKa = 4.19PGFDD219 pKa = 3.36KK220 pKa = 11.15TKK222 pKa = 9.65VAVYY226 pKa = 10.65GLGLDD231 pKa = 3.96GGSGGGNGQTQWSMYY246 pKa = 10.39SGTTGWTVTDD256 pKa = 3.85KK257 pKa = 11.44NPWGTKK263 pKa = 8.62YY264 pKa = 10.87NYY266 pKa = 10.04DD267 pKa = 3.35AAEE270 pKa = 4.31FKK272 pKa = 10.43DD273 pKa = 4.06TISWMASLAEE283 pKa = 4.25KK284 pKa = 10.67GYY286 pKa = 8.86MPTVEE291 pKa = 4.74AVTGQSSGDD300 pKa = 3.03IFGAGKK306 pKa = 10.33YY307 pKa = 11.05AMITNGSWMINQMFGYY323 pKa = 8.87TGIEE327 pKa = 3.98TGLAPTPVGPSGEE340 pKa = 4.08RR341 pKa = 11.84ASMYY345 pKa = 10.68NGLADD350 pKa = 4.14SVWAGSDD357 pKa = 3.04NKK359 pKa = 10.48AAAVKK364 pKa = 8.48WVEE367 pKa = 4.08YY368 pKa = 10.1LGSAACQDD376 pKa = 3.52IVGEE380 pKa = 4.13AGVVFPAIPSATEE393 pKa = 3.39KK394 pKa = 10.79AQAAFADD401 pKa = 3.92RR402 pKa = 11.84GIDD405 pKa = 3.46VEE407 pKa = 4.28PFLVHH412 pKa = 6.36VNEE415 pKa = 4.25GTTFLFPITDD425 pKa = 3.58YY426 pKa = 11.42ASQINGIMQPAVDD439 pKa = 4.79AVFTGQADD447 pKa = 3.85VDD449 pKa = 4.26SLDD452 pKa = 3.74AANEE456 pKa = 3.97QVNALFNGG464 pKa = 3.79
Molecular weight: 49.17 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1C0AR08|A0A1C0AR08_9ACTN Tyrosine recombinase XerD OS=Tessaracoccus lapidicaptus OX=1427523 GN=xerD PE=3 SV=1
MM1 pKa = 7.49RR2 pKa = 11.84AAPARR7 pKa = 11.84AEE9 pKa = 4.3TIDD12 pKa = 3.31GRR14 pKa = 11.84GAMMTVAVRR23 pKa = 11.84VRR25 pKa = 11.84RR26 pKa = 11.84VSVARR31 pKa = 11.84GVTGTIAVAPARR43 pKa = 11.84AVMMTVVVRR52 pKa = 11.84VRR54 pKa = 11.84RR55 pKa = 11.84VSVARR60 pKa = 11.84GATAMTIVVPVRR72 pKa = 11.84VVMMTVVVRR81 pKa = 11.84VRR83 pKa = 11.84RR84 pKa = 11.84VSVARR89 pKa = 11.84GATAMTIVAPVRR101 pKa = 11.84AVMMTVVVRR110 pKa = 11.84VRR112 pKa = 11.84RR113 pKa = 11.84VSVARR118 pKa = 11.84GASVTTTVAPALVRR132 pKa = 11.84RR133 pKa = 11.84VSVARR138 pKa = 11.84GVTGTTTVAPALVRR152 pKa = 11.84RR153 pKa = 11.84VSVARR158 pKa = 11.84GASVTTAVAPARR170 pKa = 11.84VRR172 pKa = 11.84TASVARR178 pKa = 11.84GATATIAVVPAPVVMMTGVVRR199 pKa = 11.84VRR201 pKa = 11.84RR202 pKa = 11.84VSVVRR207 pKa = 11.84GVTATIAVDD216 pKa = 3.7LVPAVMMTGVVRR228 pKa = 11.84VRR230 pKa = 11.84RR231 pKa = 11.84VSVVRR236 pKa = 11.84GVTATIAVDD245 pKa = 3.7LVPAVMMTGVARR257 pKa = 11.84VRR259 pKa = 11.84RR260 pKa = 11.84ASVVRR265 pKa = 11.84GVTATTTVDD274 pKa = 3.72LVPAVTMTAEE284 pKa = 3.78ARR286 pKa = 11.84AATMTGAVRR295 pKa = 11.84VRR297 pKa = 11.84RR298 pKa = 11.84VSVAPGATATTTVAPARR315 pKa = 11.84VATTTAEE322 pKa = 3.98ARR324 pKa = 11.84AGTTGVRR331 pKa = 11.84APRR334 pKa = 11.84NSPTSRR340 pKa = 11.84IPAPPPARR348 pKa = 11.84TSPTCRR354 pKa = 11.84RR355 pKa = 11.84TSSSPSSRR363 pKa = 11.84RR364 pKa = 11.84AA365 pKa = 3.01
MM1 pKa = 7.49RR2 pKa = 11.84AAPARR7 pKa = 11.84AEE9 pKa = 4.3TIDD12 pKa = 3.31GRR14 pKa = 11.84GAMMTVAVRR23 pKa = 11.84VRR25 pKa = 11.84RR26 pKa = 11.84VSVARR31 pKa = 11.84GVTGTIAVAPARR43 pKa = 11.84AVMMTVVVRR52 pKa = 11.84VRR54 pKa = 11.84RR55 pKa = 11.84VSVARR60 pKa = 11.84GATAMTIVVPVRR72 pKa = 11.84VVMMTVVVRR81 pKa = 11.84VRR83 pKa = 11.84RR84 pKa = 11.84VSVARR89 pKa = 11.84GATAMTIVAPVRR101 pKa = 11.84AVMMTVVVRR110 pKa = 11.84VRR112 pKa = 11.84RR113 pKa = 11.84VSVARR118 pKa = 11.84GASVTTTVAPALVRR132 pKa = 11.84RR133 pKa = 11.84VSVARR138 pKa = 11.84GVTGTTTVAPALVRR152 pKa = 11.84RR153 pKa = 11.84VSVARR158 pKa = 11.84GASVTTAVAPARR170 pKa = 11.84VRR172 pKa = 11.84TASVARR178 pKa = 11.84GATATIAVVPAPVVMMTGVVRR199 pKa = 11.84VRR201 pKa = 11.84RR202 pKa = 11.84VSVVRR207 pKa = 11.84GVTATIAVDD216 pKa = 3.7LVPAVMMTGVVRR228 pKa = 11.84VRR230 pKa = 11.84RR231 pKa = 11.84VSVVRR236 pKa = 11.84GVTATIAVDD245 pKa = 3.7LVPAVMMTGVARR257 pKa = 11.84VRR259 pKa = 11.84RR260 pKa = 11.84ASVVRR265 pKa = 11.84GVTATTTVDD274 pKa = 3.72LVPAVTMTAEE284 pKa = 3.78ARR286 pKa = 11.84AATMTGAVRR295 pKa = 11.84VRR297 pKa = 11.84RR298 pKa = 11.84VSVAPGATATTTVAPARR315 pKa = 11.84VATTTAEE322 pKa = 3.98ARR324 pKa = 11.84AGTTGVRR331 pKa = 11.84APRR334 pKa = 11.84NSPTSRR340 pKa = 11.84IPAPPPARR348 pKa = 11.84TSPTCRR354 pKa = 11.84RR355 pKa = 11.84TSSSPSSRR363 pKa = 11.84RR364 pKa = 11.84AA365 pKa = 3.01
Molecular weight: 37.51 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
921061 |
37 |
2146 |
336.6 |
36.12 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.323 ± 0.068 | 0.653 ± 0.014 |
6.385 ± 0.036 | 5.694 ± 0.043 |
2.949 ± 0.027 | 8.98 ± 0.04 |
2.172 ± 0.024 | 4.056 ± 0.031 |
1.946 ± 0.031 | 10.346 ± 0.048 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.062 ± 0.021 | 1.89 ± 0.026 |
5.48 ± 0.033 | 2.774 ± 0.026 |
7.495 ± 0.057 | 5.111 ± 0.026 |
5.985 ± 0.036 | 9.141 ± 0.044 |
1.547 ± 0.019 | 2.01 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
