
Streptomyces sp. Ag109_G2-1
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Streptomycetales; Streptomycetaceae; Streptomyces; unclassified Streptomyces
Average proteome isoelectric point is 6.51
Get precalculated fractions of proteins
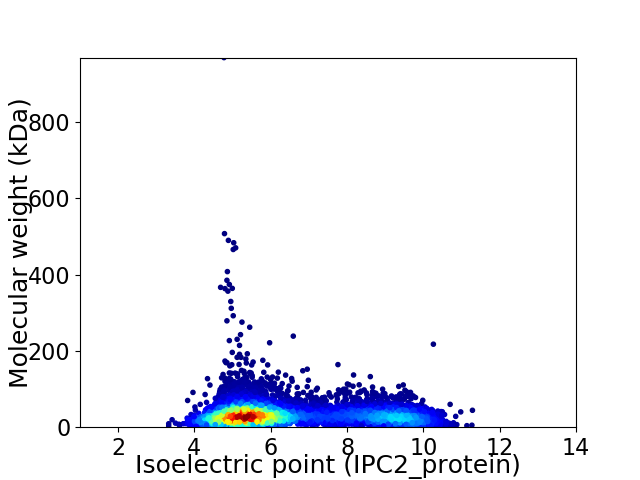
Virtual 2D-PAGE plot for 7108 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2N0FNK4|A0A2N0FNK4_9ACTN XRE family transcriptional regulator OS=Streptomyces sp. Ag109_G2-1 OX=1938849 GN=BX280_6914 PE=4 SV=1
MM1 pKa = 6.4QQEE4 pKa = 4.51VPAGGSGGAGEE15 pKa = 4.59PLEE18 pKa = 4.2VWIDD22 pKa = 3.44QDD24 pKa = 3.99LCTGDD29 pKa = 4.65GICVQYY35 pKa = 10.96APEE38 pKa = 4.18VFEE41 pKa = 5.9LDD43 pKa = 3.05IDD45 pKa = 3.63GLAYY49 pKa = 10.45VKK51 pKa = 10.83SPEE54 pKa = 4.4DD55 pKa = 3.55EE56 pKa = 4.47LLVEE60 pKa = 5.2PGATTPVPLPLLRR73 pKa = 11.84DD74 pKa = 3.74VVDD77 pKa = 4.03SAKK80 pKa = 10.21EE81 pKa = 3.92CPGDD85 pKa = 4.18CIHH88 pKa = 6.36VRR90 pKa = 11.84RR91 pKa = 11.84VSDD94 pKa = 3.3RR95 pKa = 11.84VEE97 pKa = 4.16VYY99 pKa = 10.89GPDD102 pKa = 3.45AEE104 pKa = 4.31
MM1 pKa = 6.4QQEE4 pKa = 4.51VPAGGSGGAGEE15 pKa = 4.59PLEE18 pKa = 4.2VWIDD22 pKa = 3.44QDD24 pKa = 3.99LCTGDD29 pKa = 4.65GICVQYY35 pKa = 10.96APEE38 pKa = 4.18VFEE41 pKa = 5.9LDD43 pKa = 3.05IDD45 pKa = 3.63GLAYY49 pKa = 10.45VKK51 pKa = 10.83SPEE54 pKa = 4.4DD55 pKa = 3.55EE56 pKa = 4.47LLVEE60 pKa = 5.2PGATTPVPLPLLRR73 pKa = 11.84DD74 pKa = 3.74VVDD77 pKa = 4.03SAKK80 pKa = 10.21EE81 pKa = 3.92CPGDD85 pKa = 4.18CIHH88 pKa = 6.36VRR90 pKa = 11.84RR91 pKa = 11.84VSDD94 pKa = 3.3RR95 pKa = 11.84VEE97 pKa = 4.16VYY99 pKa = 10.89GPDD102 pKa = 3.45AEE104 pKa = 4.31
Molecular weight: 11.11 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2N0H2K0|A0A2N0H2K0_9ACTN Secreted protein OS=Streptomyces sp. Ag109_G2-1 OX=1938849 GN=BX280_5548 PE=4 SV=1
MM1 pKa = 6.82VASAATTAVVAPPVAASAVTTAADD25 pKa = 3.34VRR27 pKa = 11.84PAVASVAVTTAAATVRR43 pKa = 11.84RR44 pKa = 11.84TPAVMTSAVVSVAVAMTVVGTVRR67 pKa = 11.84RR68 pKa = 11.84TRR70 pKa = 11.84VVMTAVATVPLTRR83 pKa = 11.84VVMTSVVVSVAVAMTVAATARR104 pKa = 11.84PSVVTTGAATVRR116 pKa = 11.84SRR118 pKa = 11.84RR119 pKa = 11.84VVTTTVVVSVAVVTTVVGTVRR140 pKa = 11.84RR141 pKa = 11.84TRR143 pKa = 11.84VVMTVGTVPLTRR155 pKa = 11.84VVMTSVVVSVAVVTTVVATVRR176 pKa = 11.84RR177 pKa = 11.84TRR179 pKa = 11.84VVMTAVATVRR189 pKa = 11.84PTPVVTTSVVVSVAVVTTAVGTVRR213 pKa = 11.84PTPVVMTSAVVSVATVRR230 pKa = 11.84PSVVTTGVASVPPSVVMTVGTVLPTRR256 pKa = 11.84VVTMSVAVSVAVVTTVAATVRR277 pKa = 11.84RR278 pKa = 11.84TRR280 pKa = 11.84VVTTAVATVPPTPVVMTSVVGSVVATVPPSAVTSGAATVRR320 pKa = 11.84SRR322 pKa = 11.84RR323 pKa = 11.84VGTTTVAGVLPTRR336 pKa = 11.84VVMTSAAVSVAAVTTAVATVRR357 pKa = 11.84PTGVVTTVVGTVPRR371 pKa = 11.84TAAVTTGVTVRR382 pKa = 11.84PTRR385 pKa = 11.84VVTTGAASVVATVVAAMTAVVSVAVARR412 pKa = 11.84SGAAVSSVPTATGTRR427 pKa = 11.84SSGFRR432 pKa = 11.84STTTSPATRR441 pKa = 11.84STPMCARR448 pKa = 11.84SSS450 pKa = 3.42
MM1 pKa = 6.82VASAATTAVVAPPVAASAVTTAADD25 pKa = 3.34VRR27 pKa = 11.84PAVASVAVTTAAATVRR43 pKa = 11.84RR44 pKa = 11.84TPAVMTSAVVSVAVAMTVVGTVRR67 pKa = 11.84RR68 pKa = 11.84TRR70 pKa = 11.84VVMTAVATVPLTRR83 pKa = 11.84VVMTSVVVSVAVAMTVAATARR104 pKa = 11.84PSVVTTGAATVRR116 pKa = 11.84SRR118 pKa = 11.84RR119 pKa = 11.84VVTTTVVVSVAVVTTVVGTVRR140 pKa = 11.84RR141 pKa = 11.84TRR143 pKa = 11.84VVMTVGTVPLTRR155 pKa = 11.84VVMTSVVVSVAVVTTVVATVRR176 pKa = 11.84RR177 pKa = 11.84TRR179 pKa = 11.84VVMTAVATVRR189 pKa = 11.84PTPVVTTSVVVSVAVVTTAVGTVRR213 pKa = 11.84PTPVVMTSAVVSVATVRR230 pKa = 11.84PSVVTTGVASVPPSVVMTVGTVLPTRR256 pKa = 11.84VVTMSVAVSVAVVTTVAATVRR277 pKa = 11.84RR278 pKa = 11.84TRR280 pKa = 11.84VVTTAVATVPPTPVVMTSVVGSVVATVPPSAVTSGAATVRR320 pKa = 11.84SRR322 pKa = 11.84RR323 pKa = 11.84VGTTTVAGVLPTRR336 pKa = 11.84VVMTSAAVSVAAVTTAVATVRR357 pKa = 11.84PTGVVTTVVGTVPRR371 pKa = 11.84TAAVTTGVTVRR382 pKa = 11.84PTRR385 pKa = 11.84VVTTGAASVVATVVAAMTAVVSVAVARR412 pKa = 11.84SGAAVSSVPTATGTRR427 pKa = 11.84SSGFRR432 pKa = 11.84STTTSPATRR441 pKa = 11.84STPMCARR448 pKa = 11.84SSS450 pKa = 3.42
Molecular weight: 44.33 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2357120 |
27 |
9406 |
331.6 |
35.3 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.476 ± 0.059 | 0.818 ± 0.008 |
5.609 ± 0.022 | 5.695 ± 0.027 |
2.658 ± 0.017 | 9.821 ± 0.036 |
2.274 ± 0.014 | 2.911 ± 0.025 |
2.066 ± 0.027 | 10.419 ± 0.036 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.682 ± 0.012 | 1.667 ± 0.017 |
6.533 ± 0.033 | 2.715 ± 0.017 |
8.066 ± 0.036 | 4.887 ± 0.021 |
5.918 ± 0.029 | 8.225 ± 0.033 |
1.528 ± 0.013 | 2.032 ± 0.015 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
