
Thioclava arenosa
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Rhodobacteraceae; Thioclava
Average proteome isoelectric point is 6.17
Get precalculated fractions of proteins
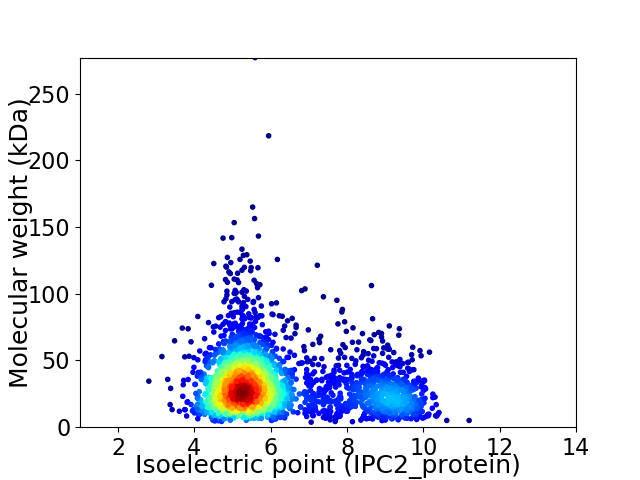
Virtual 2D-PAGE plot for 2673 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
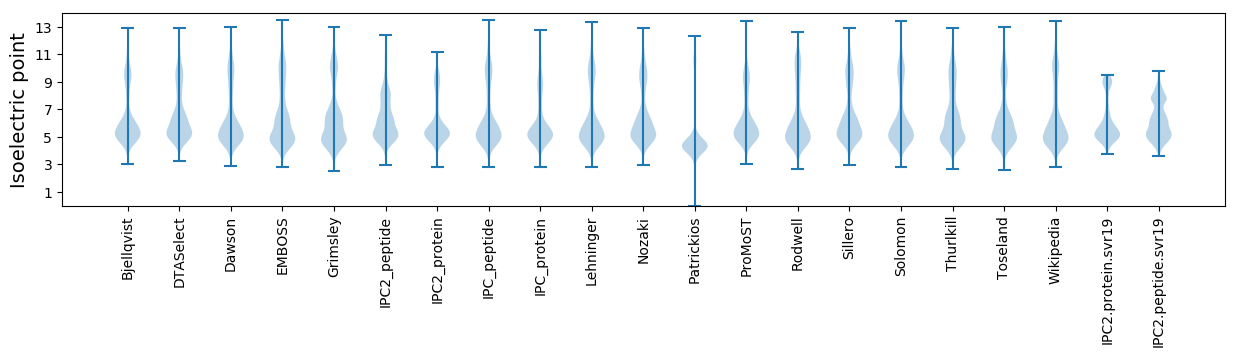
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2A4CK52|A0A2A4CK52_9RHOB Proline iminopeptidase OS=Thioclava arenosa OX=1795308 GN=pip PE=3 SV=1
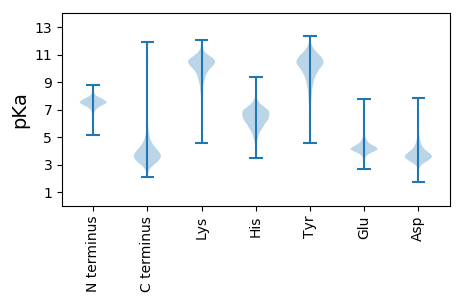
MM1 pKa = 7.22IVAAVAGCSGSAVIKK16 pKa = 10.98SGDD19 pKa = 3.55DD20 pKa = 3.13TGGGDD25 pKa = 4.94IITLPGDD32 pKa = 3.6TTSDD36 pKa = 3.38GSVSRR41 pKa = 11.84YY42 pKa = 8.49EE43 pKa = 4.32PEE45 pKa = 4.26DD46 pKa = 3.47AASGSGYY53 pKa = 10.25AQAVEE58 pKa = 4.17VRR60 pKa = 11.84DD61 pKa = 3.98NGDD64 pKa = 3.05GTTTFLVDD72 pKa = 3.27NLAFDD77 pKa = 3.78GAEE80 pKa = 3.98YY81 pKa = 11.05SEE83 pKa = 4.39GTVVSSLGRR92 pKa = 11.84TGINAGPFSVYY103 pKa = 9.9EE104 pKa = 4.27GPEE107 pKa = 3.93FEE109 pKa = 5.02PDD111 pKa = 3.64FVTGTAIAQLTHH123 pKa = 6.32RR124 pKa = 11.84AIYY127 pKa = 10.21AKK129 pKa = 9.13STSGNSEE136 pKa = 3.72FAIVRR141 pKa = 11.84TGSYY145 pKa = 8.82TGYY148 pKa = 10.76GFGGFLYY155 pKa = 10.61SRR157 pKa = 11.84TGSVTLPATATGTGISRR174 pKa = 11.84QASYY178 pKa = 11.05SGDD181 pKa = 3.47YY182 pKa = 10.89AGLIDD187 pKa = 3.79YY188 pKa = 10.02NGRR191 pKa = 11.84SGLDD195 pKa = 3.43YY196 pKa = 10.7VTGTASIAIDD206 pKa = 3.75FADD209 pKa = 4.51FDD211 pKa = 5.01DD212 pKa = 4.93GAAIYY217 pKa = 10.39GSISDD222 pKa = 4.14RR223 pKa = 11.84RR224 pKa = 11.84MFDD227 pKa = 2.66IDD229 pKa = 4.67GNDD232 pKa = 2.98ISQDD236 pKa = 3.33YY237 pKa = 10.87LDD239 pKa = 4.78RR240 pKa = 11.84LNEE243 pKa = 4.03EE244 pKa = 4.17LDD246 pKa = 3.76YY247 pKa = 10.73PAGSGYY253 pKa = 9.43TSLPTLVFDD262 pKa = 4.79IGPGVLADD270 pKa = 3.49NGEE273 pKa = 4.18IEE275 pKa = 4.41GTLGVDD281 pKa = 3.34VLTAGGVVQEE291 pKa = 4.44LVSGKK296 pKa = 10.15YY297 pKa = 9.87YY298 pKa = 10.99GVISGDD304 pKa = 3.35VTAGTDD310 pKa = 3.36EE311 pKa = 4.3VVGVIVVTGDD321 pKa = 3.1DD322 pKa = 3.58PGYY325 pKa = 10.86DD326 pKa = 3.15GVTMRR331 pKa = 11.84EE332 pKa = 3.6TGGFIVYY339 pKa = 9.79RR340 pKa = 11.84QQ341 pKa = 3.16
MM1 pKa = 7.22IVAAVAGCSGSAVIKK16 pKa = 10.98SGDD19 pKa = 3.55DD20 pKa = 3.13TGGGDD25 pKa = 4.94IITLPGDD32 pKa = 3.6TTSDD36 pKa = 3.38GSVSRR41 pKa = 11.84YY42 pKa = 8.49EE43 pKa = 4.32PEE45 pKa = 4.26DD46 pKa = 3.47AASGSGYY53 pKa = 10.25AQAVEE58 pKa = 4.17VRR60 pKa = 11.84DD61 pKa = 3.98NGDD64 pKa = 3.05GTTTFLVDD72 pKa = 3.27NLAFDD77 pKa = 3.78GAEE80 pKa = 3.98YY81 pKa = 11.05SEE83 pKa = 4.39GTVVSSLGRR92 pKa = 11.84TGINAGPFSVYY103 pKa = 9.9EE104 pKa = 4.27GPEE107 pKa = 3.93FEE109 pKa = 5.02PDD111 pKa = 3.64FVTGTAIAQLTHH123 pKa = 6.32RR124 pKa = 11.84AIYY127 pKa = 10.21AKK129 pKa = 9.13STSGNSEE136 pKa = 3.72FAIVRR141 pKa = 11.84TGSYY145 pKa = 8.82TGYY148 pKa = 10.76GFGGFLYY155 pKa = 10.61SRR157 pKa = 11.84TGSVTLPATATGTGISRR174 pKa = 11.84QASYY178 pKa = 11.05SGDD181 pKa = 3.47YY182 pKa = 10.89AGLIDD187 pKa = 3.79YY188 pKa = 10.02NGRR191 pKa = 11.84SGLDD195 pKa = 3.43YY196 pKa = 10.7VTGTASIAIDD206 pKa = 3.75FADD209 pKa = 4.51FDD211 pKa = 5.01DD212 pKa = 4.93GAAIYY217 pKa = 10.39GSISDD222 pKa = 4.14RR223 pKa = 11.84RR224 pKa = 11.84MFDD227 pKa = 2.66IDD229 pKa = 4.67GNDD232 pKa = 2.98ISQDD236 pKa = 3.33YY237 pKa = 10.87LDD239 pKa = 4.78RR240 pKa = 11.84LNEE243 pKa = 4.03EE244 pKa = 4.17LDD246 pKa = 3.76YY247 pKa = 10.73PAGSGYY253 pKa = 9.43TSLPTLVFDD262 pKa = 4.79IGPGVLADD270 pKa = 3.49NGEE273 pKa = 4.18IEE275 pKa = 4.41GTLGVDD281 pKa = 3.34VLTAGGVVQEE291 pKa = 4.44LVSGKK296 pKa = 10.15YY297 pKa = 9.87YY298 pKa = 10.99GVISGDD304 pKa = 3.35VTAGTDD310 pKa = 3.36EE311 pKa = 4.3VVGVIVVTGDD321 pKa = 3.1DD322 pKa = 3.58PGYY325 pKa = 10.86DD326 pKa = 3.15GVTMRR331 pKa = 11.84EE332 pKa = 3.6TGGFIVYY339 pKa = 9.79RR340 pKa = 11.84QQ341 pKa = 3.16
Molecular weight: 35.09 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2A4CP57|A0A2A4CP57_9RHOB Periplasmic nitrate reductase OS=Thioclava arenosa OX=1795308 GN=napA PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84ARR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.37GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.42GGQKK29 pKa = 9.85VLNARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.69GRR39 pKa = 11.84KK40 pKa = 8.91RR41 pKa = 11.84LSAA44 pKa = 3.96
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84ARR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.37GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.42GGQKK29 pKa = 9.85VLNARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.69GRR39 pKa = 11.84KK40 pKa = 8.91RR41 pKa = 11.84LSAA44 pKa = 3.96
Molecular weight: 5.07 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
840682 |
37 |
2492 |
314.5 |
34.05 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.208 ± 0.079 | 0.857 ± 0.015 |
5.514 ± 0.036 | 6.477 ± 0.047 |
3.685 ± 0.026 | 8.878 ± 0.048 |
1.971 ± 0.02 | 5.074 ± 0.03 |
3.198 ± 0.041 | 10.401 ± 0.064 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.744 ± 0.023 | 2.334 ± 0.023 |
5.326 ± 0.041 | 2.991 ± 0.023 |
6.942 ± 0.048 | 4.783 ± 0.032 |
5.032 ± 0.035 | 7.088 ± 0.037 |
1.372 ± 0.021 | 2.125 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
