
Imperata yellow mottle virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Pisoniviricetes; Sobelivirales; Solemoviridae; Sobemovirus
Average proteome isoelectric point is 7.32
Get precalculated fractions of proteins
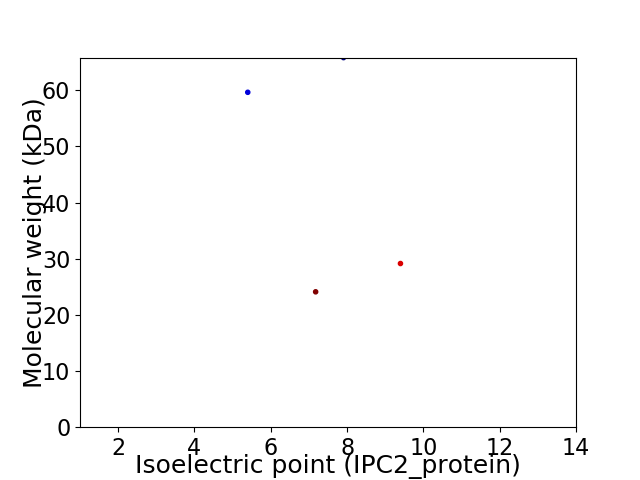
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|B5WYM8|B5WYM8_9VIRU Capsid protein OS=Imperata yellow mottle virus OX=524023 PE=3 SV=1
MM1 pKa = 7.92PLTAVGRR8 pKa = 11.84SDD10 pKa = 3.13CRR12 pKa = 11.84FRR14 pKa = 11.84EE15 pKa = 4.93GTQKK19 pKa = 10.75PPGPRR24 pKa = 11.84VSAAVRR30 pKa = 11.84VFPEE34 pKa = 4.01LADD37 pKa = 3.87YY38 pKa = 10.15AWPARR43 pKa = 11.84GSEE46 pKa = 4.38AEE48 pKa = 4.19LEE50 pKa = 4.32SLRR53 pKa = 11.84VQASRR58 pKa = 11.84FKK60 pKa = 10.32RR61 pKa = 11.84TLPPPSLQEE70 pKa = 3.99AVNALQGRR78 pKa = 11.84YY79 pKa = 9.15PNSQVRR85 pKa = 11.84KK86 pKa = 9.6CFRR89 pKa = 11.84QEE91 pKa = 3.36PWDD94 pKa = 3.95FQALCEE100 pKa = 4.01EE101 pKa = 4.55VQRR104 pKa = 11.84ICQTGEE110 pKa = 3.87LNGTSSPGVPLANIEE125 pKa = 4.19NQNAAVLNLAPEE137 pKa = 4.53LVHH140 pKa = 6.71LAVAEE145 pKa = 4.06RR146 pKa = 11.84LVALAAVDD154 pKa = 3.88PRR156 pKa = 11.84QHH158 pKa = 5.41NWTPKK163 pKa = 9.52EE164 pKa = 3.76LVQRR168 pKa = 11.84GLVDD172 pKa = 3.62PVRR175 pKa = 11.84LFVKK179 pKa = 10.12QEE181 pKa = 3.57PHH183 pKa = 4.42TMRR186 pKa = 11.84KK187 pKa = 8.0IRR189 pKa = 11.84EE190 pKa = 3.7RR191 pKa = 11.84RR192 pKa = 11.84FRR194 pKa = 11.84LISSVSLVDD203 pKa = 3.53QLVEE207 pKa = 4.17RR208 pKa = 11.84MLFGPQNATEE218 pKa = 3.6ISMWHH223 pKa = 6.65LCPSKK228 pKa = 10.38PGMGMSTPSQVEE240 pKa = 4.18MLWKK244 pKa = 10.5DD245 pKa = 3.58VAHH248 pKa = 6.47KK249 pKa = 10.46HH250 pKa = 5.59SLHH253 pKa = 5.74QAAEE257 pKa = 4.0ADD259 pKa = 3.05ISAFDD264 pKa = 3.67WSVQDD269 pKa = 3.38WEE271 pKa = 4.52LWADD275 pKa = 3.73LAIRR279 pKa = 11.84LQLGSFPDD287 pKa = 3.67LMRR290 pKa = 11.84RR291 pKa = 11.84AAISRR296 pKa = 11.84FYY298 pKa = 11.3CFMNSVFQLSDD309 pKa = 3.11GTLIEE314 pKa = 4.14QNLPGLMKK322 pKa = 10.49SGSYY326 pKa = 8.59CTSSTNSRR334 pKa = 11.84IRR336 pKa = 11.84CLMAEE341 pKa = 4.85LIGSPWCIAMGDD353 pKa = 3.8DD354 pKa = 4.68SVEE357 pKa = 3.99GWVEE361 pKa = 4.15GAQEE365 pKa = 3.91KK366 pKa = 10.09YY367 pKa = 10.39AALGHH372 pKa = 5.17TCKK375 pKa = 10.4EE376 pKa = 4.28YY377 pKa = 10.75VACPSKK383 pKa = 10.5QGRR386 pKa = 11.84LGKK389 pKa = 10.11EE390 pKa = 3.29LLSFNFCSHH399 pKa = 6.38EE400 pKa = 4.01FTNPGLPRR408 pKa = 11.84AEE410 pKa = 4.04LLTWAKK416 pKa = 10.1CLYY419 pKa = 10.38RR420 pKa = 11.84FLSSNRR426 pKa = 11.84EE427 pKa = 4.01TVDD430 pKa = 3.96DD431 pKa = 4.67LWVEE435 pKa = 4.52LNTSRR440 pKa = 11.84QWGRR444 pKa = 11.84IQKK447 pKa = 8.37YY448 pKa = 8.9LAGIGEE454 pKa = 4.27VSQLNSADD462 pKa = 3.77GEE464 pKa = 4.35IKK466 pKa = 10.72AEE468 pKa = 4.33EE469 pKa = 4.13IQEE472 pKa = 4.18DD473 pKa = 3.81AAGCGGQEE481 pKa = 3.9PAASAFEE488 pKa = 4.04EE489 pKa = 4.23ARR491 pKa = 11.84EE492 pKa = 3.94IWIEE496 pKa = 3.86PTPAQQWDD504 pKa = 4.2NNIGPHH510 pKa = 5.68GYY512 pKa = 9.2EE513 pKa = 3.73WDD515 pKa = 3.66WPFSEE520 pKa = 5.26SWSPWGRR527 pKa = 11.84DD528 pKa = 3.16HH529 pKa = 7.69SS530 pKa = 4.09
MM1 pKa = 7.92PLTAVGRR8 pKa = 11.84SDD10 pKa = 3.13CRR12 pKa = 11.84FRR14 pKa = 11.84EE15 pKa = 4.93GTQKK19 pKa = 10.75PPGPRR24 pKa = 11.84VSAAVRR30 pKa = 11.84VFPEE34 pKa = 4.01LADD37 pKa = 3.87YY38 pKa = 10.15AWPARR43 pKa = 11.84GSEE46 pKa = 4.38AEE48 pKa = 4.19LEE50 pKa = 4.32SLRR53 pKa = 11.84VQASRR58 pKa = 11.84FKK60 pKa = 10.32RR61 pKa = 11.84TLPPPSLQEE70 pKa = 3.99AVNALQGRR78 pKa = 11.84YY79 pKa = 9.15PNSQVRR85 pKa = 11.84KK86 pKa = 9.6CFRR89 pKa = 11.84QEE91 pKa = 3.36PWDD94 pKa = 3.95FQALCEE100 pKa = 4.01EE101 pKa = 4.55VQRR104 pKa = 11.84ICQTGEE110 pKa = 3.87LNGTSSPGVPLANIEE125 pKa = 4.19NQNAAVLNLAPEE137 pKa = 4.53LVHH140 pKa = 6.71LAVAEE145 pKa = 4.06RR146 pKa = 11.84LVALAAVDD154 pKa = 3.88PRR156 pKa = 11.84QHH158 pKa = 5.41NWTPKK163 pKa = 9.52EE164 pKa = 3.76LVQRR168 pKa = 11.84GLVDD172 pKa = 3.62PVRR175 pKa = 11.84LFVKK179 pKa = 10.12QEE181 pKa = 3.57PHH183 pKa = 4.42TMRR186 pKa = 11.84KK187 pKa = 8.0IRR189 pKa = 11.84EE190 pKa = 3.7RR191 pKa = 11.84RR192 pKa = 11.84FRR194 pKa = 11.84LISSVSLVDD203 pKa = 3.53QLVEE207 pKa = 4.17RR208 pKa = 11.84MLFGPQNATEE218 pKa = 3.6ISMWHH223 pKa = 6.65LCPSKK228 pKa = 10.38PGMGMSTPSQVEE240 pKa = 4.18MLWKK244 pKa = 10.5DD245 pKa = 3.58VAHH248 pKa = 6.47KK249 pKa = 10.46HH250 pKa = 5.59SLHH253 pKa = 5.74QAAEE257 pKa = 4.0ADD259 pKa = 3.05ISAFDD264 pKa = 3.67WSVQDD269 pKa = 3.38WEE271 pKa = 4.52LWADD275 pKa = 3.73LAIRR279 pKa = 11.84LQLGSFPDD287 pKa = 3.67LMRR290 pKa = 11.84RR291 pKa = 11.84AAISRR296 pKa = 11.84FYY298 pKa = 11.3CFMNSVFQLSDD309 pKa = 3.11GTLIEE314 pKa = 4.14QNLPGLMKK322 pKa = 10.49SGSYY326 pKa = 8.59CTSSTNSRR334 pKa = 11.84IRR336 pKa = 11.84CLMAEE341 pKa = 4.85LIGSPWCIAMGDD353 pKa = 3.8DD354 pKa = 4.68SVEE357 pKa = 3.99GWVEE361 pKa = 4.15GAQEE365 pKa = 3.91KK366 pKa = 10.09YY367 pKa = 10.39AALGHH372 pKa = 5.17TCKK375 pKa = 10.4EE376 pKa = 4.28YY377 pKa = 10.75VACPSKK383 pKa = 10.5QGRR386 pKa = 11.84LGKK389 pKa = 10.11EE390 pKa = 3.29LLSFNFCSHH399 pKa = 6.38EE400 pKa = 4.01FTNPGLPRR408 pKa = 11.84AEE410 pKa = 4.04LLTWAKK416 pKa = 10.1CLYY419 pKa = 10.38RR420 pKa = 11.84FLSSNRR426 pKa = 11.84EE427 pKa = 4.01TVDD430 pKa = 3.96DD431 pKa = 4.67LWVEE435 pKa = 4.52LNTSRR440 pKa = 11.84QWGRR444 pKa = 11.84IQKK447 pKa = 8.37YY448 pKa = 8.9LAGIGEE454 pKa = 4.27VSQLNSADD462 pKa = 3.77GEE464 pKa = 4.35IKK466 pKa = 10.72AEE468 pKa = 4.33EE469 pKa = 4.13IQEE472 pKa = 4.18DD473 pKa = 3.81AAGCGGQEE481 pKa = 3.9PAASAFEE488 pKa = 4.04EE489 pKa = 4.23ARR491 pKa = 11.84EE492 pKa = 3.94IWIEE496 pKa = 3.86PTPAQQWDD504 pKa = 4.2NNIGPHH510 pKa = 5.68GYY512 pKa = 9.2EE513 pKa = 3.73WDD515 pKa = 3.66WPFSEE520 pKa = 5.26SWSPWGRR527 pKa = 11.84DD528 pKa = 3.16HH529 pKa = 7.69SS530 pKa = 4.09
Molecular weight: 59.63 kDa
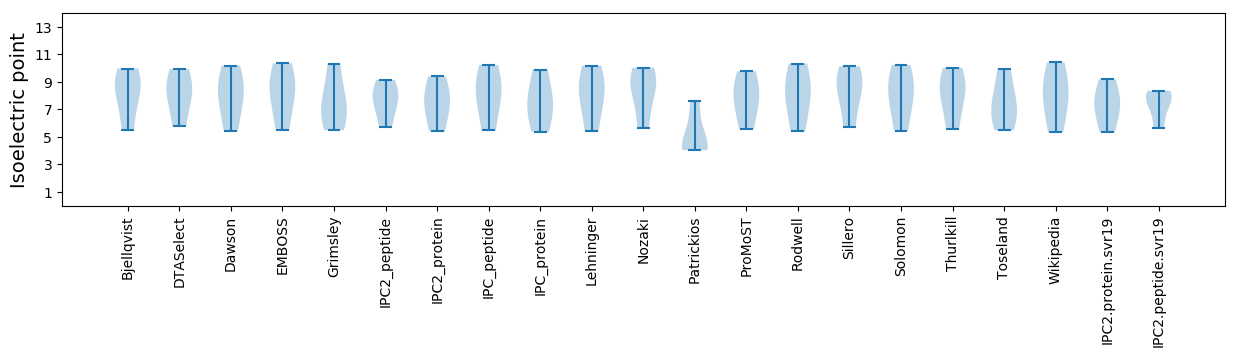
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|B5WYM8|B5WYM8_9VIRU Capsid protein OS=Imperata yellow mottle virus OX=524023 PE=3 SV=1
MM1 pKa = 7.31GRR3 pKa = 11.84SKK5 pKa = 10.63QKK7 pKa = 9.9KK8 pKa = 7.56SKK10 pKa = 10.45RR11 pKa = 11.84MPQAAAVKK19 pKa = 8.83NQQPAPSRR27 pKa = 11.84RR28 pKa = 11.84QGRR31 pKa = 11.84SGSSQRR37 pKa = 11.84QPSNGITTSAPMATSGIGPSRR58 pKa = 11.84SLGPLGGAITVEE70 pKa = 4.42EE71 pKa = 4.56VEE73 pKa = 4.31HH74 pKa = 6.83LGAFTRR80 pKa = 11.84GTTAAGQTRR89 pKa = 11.84AYY91 pKa = 10.8LCMPSSLIRR100 pKa = 11.84LSNIARR106 pKa = 11.84CFARR110 pKa = 11.84WRR112 pKa = 11.84PLQWSVFYY120 pKa = 10.85VPEE123 pKa = 4.58VGTQTNGAIQMAYY136 pKa = 10.21LYY138 pKa = 10.09DD139 pKa = 4.12YY140 pKa = 11.13GDD142 pKa = 4.2SLPTTTGQISACSGFLTAAVWCGAAGAQLLSGKK175 pKa = 10.13APRR178 pKa = 11.84SQNLVIAHH186 pKa = 5.7MDD188 pKa = 3.43CRR190 pKa = 11.84RR191 pKa = 11.84SEE193 pKa = 3.75WMRR196 pKa = 11.84VVDD199 pKa = 5.32GLTDD203 pKa = 3.41TDD205 pKa = 4.03PAHH208 pKa = 6.39VVNTYY213 pKa = 10.26LPARR217 pKa = 11.84AVVRR221 pKa = 11.84SSLLPSTEE229 pKa = 3.93DD230 pKa = 3.2TPGQLYY236 pKa = 9.44VRR238 pKa = 11.84YY239 pKa = 9.99RR240 pKa = 11.84IMLRR244 pKa = 11.84DD245 pKa = 3.52AVAPGLNDD253 pKa = 3.53ASSSAPSVDD262 pKa = 3.76QEE264 pKa = 4.43RR265 pKa = 11.84KK266 pKa = 9.36GNAAVVSS273 pKa = 4.07
MM1 pKa = 7.31GRR3 pKa = 11.84SKK5 pKa = 10.63QKK7 pKa = 9.9KK8 pKa = 7.56SKK10 pKa = 10.45RR11 pKa = 11.84MPQAAAVKK19 pKa = 8.83NQQPAPSRR27 pKa = 11.84RR28 pKa = 11.84QGRR31 pKa = 11.84SGSSQRR37 pKa = 11.84QPSNGITTSAPMATSGIGPSRR58 pKa = 11.84SLGPLGGAITVEE70 pKa = 4.42EE71 pKa = 4.56VEE73 pKa = 4.31HH74 pKa = 6.83LGAFTRR80 pKa = 11.84GTTAAGQTRR89 pKa = 11.84AYY91 pKa = 10.8LCMPSSLIRR100 pKa = 11.84LSNIARR106 pKa = 11.84CFARR110 pKa = 11.84WRR112 pKa = 11.84PLQWSVFYY120 pKa = 10.85VPEE123 pKa = 4.58VGTQTNGAIQMAYY136 pKa = 10.21LYY138 pKa = 10.09DD139 pKa = 4.12YY140 pKa = 11.13GDD142 pKa = 4.2SLPTTTGQISACSGFLTAAVWCGAAGAQLLSGKK175 pKa = 10.13APRR178 pKa = 11.84SQNLVIAHH186 pKa = 5.7MDD188 pKa = 3.43CRR190 pKa = 11.84RR191 pKa = 11.84SEE193 pKa = 3.75WMRR196 pKa = 11.84VVDD199 pKa = 5.32GLTDD203 pKa = 3.41TDD205 pKa = 4.03PAHH208 pKa = 6.39VVNTYY213 pKa = 10.26LPARR217 pKa = 11.84AVVRR221 pKa = 11.84SSLLPSTEE229 pKa = 3.93DD230 pKa = 3.2TPGQLYY236 pKa = 9.44VRR238 pKa = 11.84YY239 pKa = 9.99RR240 pKa = 11.84IMLRR244 pKa = 11.84DD245 pKa = 3.52AVAPGLNDD253 pKa = 3.53ASSSAPSVDD262 pKa = 3.76QEE264 pKa = 4.43RR265 pKa = 11.84KK266 pKa = 9.36GNAAVVSS273 pKa = 4.07
Molecular weight: 29.14 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1614 |
213 |
598 |
403.5 |
44.67 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.993 ± 1.142 | 2.23 ± 0.357 |
4.337 ± 0.319 | 6.134 ± 1.031 |
2.85 ± 0.424 | 7.249 ± 0.642 |
2.292 ± 0.406 | 3.408 ± 0.27 |
3.47 ± 0.583 | 8.488 ± 0.775 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.107 ± 0.316 | 3.346 ± 0.153 |
6.629 ± 0.241 | 4.833 ± 0.523 |
7.621 ± 0.794 | 10.223 ± 0.926 |
5.328 ± 0.765 | 6.629 ± 0.281 |
2.292 ± 0.485 | 2.54 ± 0.431 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
