
Thielaviopsis basicola mitovirus
Taxonomy: Viruses; Riboviria; Orthornavirae; Lenarviricota; Howeltoviricetes; Cryppavirales; Mitoviridae; Mitovirus; unclassified Mitovirus
Average proteome isoelectric point is 8.73
Get precalculated fractions of proteins
Virtual 2D-PAGE plot for 1 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
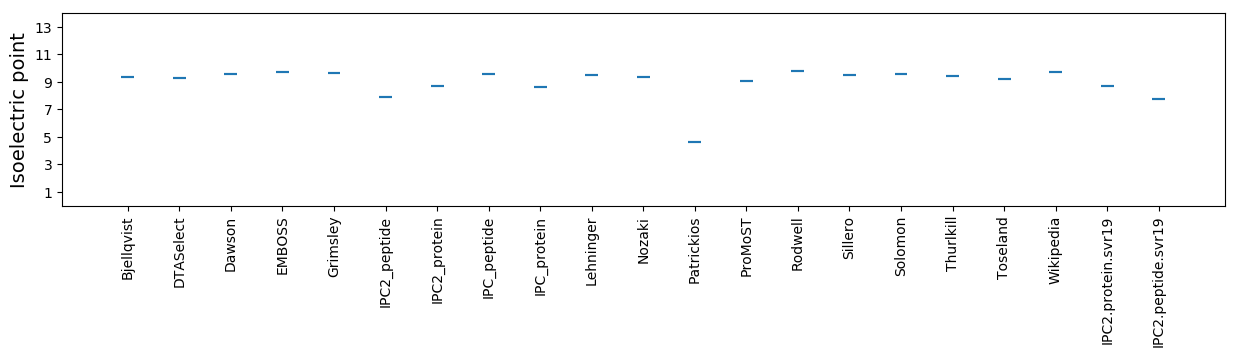
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q6Q2W5|Q6Q2W5_9VIRU RNA-dependent RNA polymerase OS=Thielaviopsis basicola mitovirus OX=274584 PE=4 SV=1
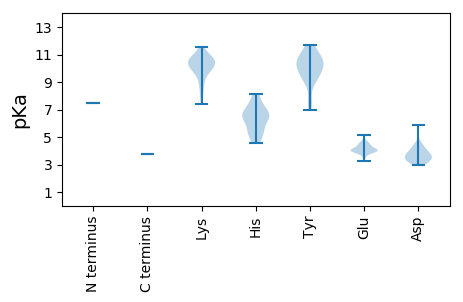
MM1 pKa = 7.44IKK3 pKa = 10.43YY4 pKa = 7.0MKK6 pKa = 9.95SVRR9 pKa = 11.84LHH11 pKa = 4.55ITRR14 pKa = 11.84YY15 pKa = 8.93ICRR18 pKa = 11.84KK19 pKa = 8.83PLFINDD25 pKa = 3.55SGVSVDD31 pKa = 4.31KK32 pKa = 11.37SGFPTKK38 pKa = 10.53FIEE41 pKa = 5.17LKK43 pKa = 10.67KK44 pKa = 10.48FLNNHH49 pKa = 5.82EE50 pKa = 4.41SSICYY55 pKa = 7.68HH56 pKa = 5.15TRR58 pKa = 11.84LRR60 pKa = 11.84IILTLLLLPRR70 pKa = 11.84GLKK73 pKa = 7.37PTKK76 pKa = 10.27IEE78 pKa = 4.01DD79 pKa = 3.55AKK81 pKa = 10.72IKK83 pKa = 10.58PNFSSITLPFSGTRR97 pKa = 11.84MNPIPTWFIKK107 pKa = 10.62EE108 pKa = 3.88FVKK111 pKa = 10.65EE112 pKa = 3.97YY113 pKa = 10.64NLKK116 pKa = 10.58LEE118 pKa = 4.4KK119 pKa = 9.22PTYY122 pKa = 10.12DD123 pKa = 3.2LSKK126 pKa = 10.58HH127 pKa = 4.92YY128 pKa = 10.12LSPKK132 pKa = 8.55GGPMGKK138 pKa = 8.08STWSSIWSMYY148 pKa = 9.59ALKK151 pKa = 10.72HH152 pKa = 5.42EE153 pKa = 4.41LHH155 pKa = 6.61LSLSYY160 pKa = 10.78FLGEE164 pKa = 3.9AYY166 pKa = 7.57KK167 pKa = 10.11TLFYY171 pKa = 11.01LPYY174 pKa = 9.15LKK176 pKa = 10.86NMALSYY182 pKa = 10.94GHH184 pKa = 7.23DD185 pKa = 3.9VPPKK189 pKa = 8.34QWPSGKK195 pKa = 10.26LGIVKK200 pKa = 10.26DD201 pKa = 4.0PEE203 pKa = 4.01GKK205 pKa = 9.82RR206 pKa = 11.84RR207 pKa = 11.84IIAMVDD213 pKa = 3.06YY214 pKa = 10.6HH215 pKa = 6.58SQLVLRR221 pKa = 11.84SIHH224 pKa = 7.51DD225 pKa = 3.56GLLNKK230 pKa = 9.71LRR232 pKa = 11.84NLPQDD237 pKa = 2.95RR238 pKa = 11.84TYY240 pKa = 11.55NQDD243 pKa = 3.38PNNAWEE249 pKa = 4.04EE250 pKa = 4.07NKK252 pKa = 10.54EE253 pKa = 4.24CFHH256 pKa = 8.14SLDD259 pKa = 4.13LSSATDD265 pKa = 3.76RR266 pKa = 11.84FPVKK270 pKa = 10.18LQSRR274 pKa = 11.84LLTEE278 pKa = 4.7MYY280 pKa = 10.32SDD282 pKa = 3.98PSFGEE287 pKa = 3.26NWMNLLLNRR296 pKa = 11.84DD297 pKa = 3.63YY298 pKa = 11.49LLPEE302 pKa = 4.46EE303 pKa = 4.59GLSGEE308 pKa = 4.16RR309 pKa = 11.84LRR311 pKa = 11.84YY312 pKa = 9.52AVGQPMGAYY321 pKa = 9.89SSWAAFTLSHH331 pKa = 6.52HH332 pKa = 6.89LVVAWCTYY340 pKa = 9.64KK341 pKa = 10.44SKK343 pKa = 11.02KK344 pKa = 9.8VIRR347 pKa = 11.84SSQYY351 pKa = 10.25IILGDD356 pKa = 4.4DD357 pKa = 3.66IVIKK361 pKa = 11.03DD362 pKa = 3.33NDD364 pKa = 3.4IARR367 pKa = 11.84KK368 pKa = 10.13YY369 pKa = 9.03IGQMSKK375 pKa = 10.84LGVAISMQKK384 pKa = 7.76THH386 pKa = 5.51VSKK389 pKa = 10.08DD390 pKa = 3.06TYY392 pKa = 10.88EE393 pKa = 3.94FAKK396 pKa = 10.15RR397 pKa = 11.84WMHH400 pKa = 6.36KK401 pKa = 9.87GVEE404 pKa = 4.09ISGLPLKK411 pKa = 10.87GIFYY415 pKa = 10.02NISHH419 pKa = 6.48MRR421 pKa = 11.84RR422 pKa = 11.84MYY424 pKa = 10.28TIIFDD429 pKa = 3.78YY430 pKa = 10.47LQRR433 pKa = 11.84IPSKK437 pKa = 11.32SNLTSLQLFASCLSGIKK454 pKa = 9.8FGKK457 pKa = 9.6RR458 pKa = 11.84VYY460 pKa = 10.88SKK462 pKa = 11.56ALILKK467 pKa = 9.19LLRR470 pKa = 11.84NFNISLRR477 pKa = 11.84YY478 pKa = 9.65SYY480 pKa = 11.56GLLTPYY486 pKa = 10.41EE487 pKa = 4.09LRR489 pKa = 11.84EE490 pKa = 4.06YY491 pKa = 10.56ILTGFNMGDD500 pKa = 3.19EE501 pKa = 4.58VLPSDD506 pKa = 4.63NRR508 pKa = 11.84ILEE511 pKa = 4.13WFKK514 pKa = 11.4GIICDD519 pKa = 3.86GVAGKK524 pKa = 10.74VSTLSRR530 pKa = 11.84EE531 pKa = 3.89YY532 pKa = 9.73TKK534 pKa = 10.64EE535 pKa = 3.99VNSLEE540 pKa = 4.16EE541 pKa = 4.22HH542 pKa = 6.85RR543 pKa = 11.84SSQHH547 pKa = 5.33ILNSINIEE555 pKa = 3.63ITYY558 pKa = 10.73EE559 pKa = 3.89NLQWSPLIIGMFNHH573 pKa = 6.38INKK576 pKa = 9.19VSEE579 pKa = 4.31KK580 pKa = 10.23CGEE583 pKa = 3.88WLYY586 pKa = 11.67GDD588 pKa = 4.34VPLMDD593 pKa = 5.88IIQQFTLPSADD604 pKa = 3.27SLSRR608 pKa = 11.84KK609 pKa = 9.92DD610 pKa = 3.48RR611 pKa = 11.84DD612 pKa = 3.35INKK615 pKa = 10.27KK616 pKa = 9.59IDD618 pKa = 3.71EE619 pKa = 4.67LDD621 pKa = 3.44SLIFKK626 pKa = 9.95SIKK629 pKa = 9.9RR630 pKa = 11.84HH631 pKa = 6.22FSDD634 pKa = 4.3QPYY637 pKa = 9.32PDD639 pKa = 3.92HH640 pKa = 7.65FYY642 pKa = 10.09GTRR645 pKa = 11.84SNGDD649 pKa = 3.06KK650 pKa = 10.77NLYY653 pKa = 9.05PGLGALDD660 pKa = 3.61VNVAPEE666 pKa = 3.64IGFEE670 pKa = 3.95IKK672 pKa = 10.76SPINFILKK680 pKa = 8.86TEE682 pKa = 4.0IQRR685 pKa = 11.84LHH687 pKa = 6.74GNLEE691 pKa = 3.99MQMEE695 pKa = 4.43KK696 pKa = 10.42PLRR699 pKa = 11.84EE700 pKa = 4.08AGIKK704 pKa = 9.87CC705 pKa = 3.76
MM1 pKa = 7.44IKK3 pKa = 10.43YY4 pKa = 7.0MKK6 pKa = 9.95SVRR9 pKa = 11.84LHH11 pKa = 4.55ITRR14 pKa = 11.84YY15 pKa = 8.93ICRR18 pKa = 11.84KK19 pKa = 8.83PLFINDD25 pKa = 3.55SGVSVDD31 pKa = 4.31KK32 pKa = 11.37SGFPTKK38 pKa = 10.53FIEE41 pKa = 5.17LKK43 pKa = 10.67KK44 pKa = 10.48FLNNHH49 pKa = 5.82EE50 pKa = 4.41SSICYY55 pKa = 7.68HH56 pKa = 5.15TRR58 pKa = 11.84LRR60 pKa = 11.84IILTLLLLPRR70 pKa = 11.84GLKK73 pKa = 7.37PTKK76 pKa = 10.27IEE78 pKa = 4.01DD79 pKa = 3.55AKK81 pKa = 10.72IKK83 pKa = 10.58PNFSSITLPFSGTRR97 pKa = 11.84MNPIPTWFIKK107 pKa = 10.62EE108 pKa = 3.88FVKK111 pKa = 10.65EE112 pKa = 3.97YY113 pKa = 10.64NLKK116 pKa = 10.58LEE118 pKa = 4.4KK119 pKa = 9.22PTYY122 pKa = 10.12DD123 pKa = 3.2LSKK126 pKa = 10.58HH127 pKa = 4.92YY128 pKa = 10.12LSPKK132 pKa = 8.55GGPMGKK138 pKa = 8.08STWSSIWSMYY148 pKa = 9.59ALKK151 pKa = 10.72HH152 pKa = 5.42EE153 pKa = 4.41LHH155 pKa = 6.61LSLSYY160 pKa = 10.78FLGEE164 pKa = 3.9AYY166 pKa = 7.57KK167 pKa = 10.11TLFYY171 pKa = 11.01LPYY174 pKa = 9.15LKK176 pKa = 10.86NMALSYY182 pKa = 10.94GHH184 pKa = 7.23DD185 pKa = 3.9VPPKK189 pKa = 8.34QWPSGKK195 pKa = 10.26LGIVKK200 pKa = 10.26DD201 pKa = 4.0PEE203 pKa = 4.01GKK205 pKa = 9.82RR206 pKa = 11.84RR207 pKa = 11.84IIAMVDD213 pKa = 3.06YY214 pKa = 10.6HH215 pKa = 6.58SQLVLRR221 pKa = 11.84SIHH224 pKa = 7.51DD225 pKa = 3.56GLLNKK230 pKa = 9.71LRR232 pKa = 11.84NLPQDD237 pKa = 2.95RR238 pKa = 11.84TYY240 pKa = 11.55NQDD243 pKa = 3.38PNNAWEE249 pKa = 4.04EE250 pKa = 4.07NKK252 pKa = 10.54EE253 pKa = 4.24CFHH256 pKa = 8.14SLDD259 pKa = 4.13LSSATDD265 pKa = 3.76RR266 pKa = 11.84FPVKK270 pKa = 10.18LQSRR274 pKa = 11.84LLTEE278 pKa = 4.7MYY280 pKa = 10.32SDD282 pKa = 3.98PSFGEE287 pKa = 3.26NWMNLLLNRR296 pKa = 11.84DD297 pKa = 3.63YY298 pKa = 11.49LLPEE302 pKa = 4.46EE303 pKa = 4.59GLSGEE308 pKa = 4.16RR309 pKa = 11.84LRR311 pKa = 11.84YY312 pKa = 9.52AVGQPMGAYY321 pKa = 9.89SSWAAFTLSHH331 pKa = 6.52HH332 pKa = 6.89LVVAWCTYY340 pKa = 9.64KK341 pKa = 10.44SKK343 pKa = 11.02KK344 pKa = 9.8VIRR347 pKa = 11.84SSQYY351 pKa = 10.25IILGDD356 pKa = 4.4DD357 pKa = 3.66IVIKK361 pKa = 11.03DD362 pKa = 3.33NDD364 pKa = 3.4IARR367 pKa = 11.84KK368 pKa = 10.13YY369 pKa = 9.03IGQMSKK375 pKa = 10.84LGVAISMQKK384 pKa = 7.76THH386 pKa = 5.51VSKK389 pKa = 10.08DD390 pKa = 3.06TYY392 pKa = 10.88EE393 pKa = 3.94FAKK396 pKa = 10.15RR397 pKa = 11.84WMHH400 pKa = 6.36KK401 pKa = 9.87GVEE404 pKa = 4.09ISGLPLKK411 pKa = 10.87GIFYY415 pKa = 10.02NISHH419 pKa = 6.48MRR421 pKa = 11.84RR422 pKa = 11.84MYY424 pKa = 10.28TIIFDD429 pKa = 3.78YY430 pKa = 10.47LQRR433 pKa = 11.84IPSKK437 pKa = 11.32SNLTSLQLFASCLSGIKK454 pKa = 9.8FGKK457 pKa = 9.6RR458 pKa = 11.84VYY460 pKa = 10.88SKK462 pKa = 11.56ALILKK467 pKa = 9.19LLRR470 pKa = 11.84NFNISLRR477 pKa = 11.84YY478 pKa = 9.65SYY480 pKa = 11.56GLLTPYY486 pKa = 10.41EE487 pKa = 4.09LRR489 pKa = 11.84EE490 pKa = 4.06YY491 pKa = 10.56ILTGFNMGDD500 pKa = 3.19EE501 pKa = 4.58VLPSDD506 pKa = 4.63NRR508 pKa = 11.84ILEE511 pKa = 4.13WFKK514 pKa = 11.4GIICDD519 pKa = 3.86GVAGKK524 pKa = 10.74VSTLSRR530 pKa = 11.84EE531 pKa = 3.89YY532 pKa = 9.73TKK534 pKa = 10.64EE535 pKa = 3.99VNSLEE540 pKa = 4.16EE541 pKa = 4.22HH542 pKa = 6.85RR543 pKa = 11.84SSQHH547 pKa = 5.33ILNSINIEE555 pKa = 3.63ITYY558 pKa = 10.73EE559 pKa = 3.89NLQWSPLIIGMFNHH573 pKa = 6.38INKK576 pKa = 9.19VSEE579 pKa = 4.31KK580 pKa = 10.23CGEE583 pKa = 3.88WLYY586 pKa = 11.67GDD588 pKa = 4.34VPLMDD593 pKa = 5.88IIQQFTLPSADD604 pKa = 3.27SLSRR608 pKa = 11.84KK609 pKa = 9.92DD610 pKa = 3.48RR611 pKa = 11.84DD612 pKa = 3.35INKK615 pKa = 10.27KK616 pKa = 9.59IDD618 pKa = 3.71EE619 pKa = 4.67LDD621 pKa = 3.44SLIFKK626 pKa = 9.95SIKK629 pKa = 9.9RR630 pKa = 11.84HH631 pKa = 6.22FSDD634 pKa = 4.3QPYY637 pKa = 9.32PDD639 pKa = 3.92HH640 pKa = 7.65FYY642 pKa = 10.09GTRR645 pKa = 11.84SNGDD649 pKa = 3.06KK650 pKa = 10.77NLYY653 pKa = 9.05PGLGALDD660 pKa = 3.61VNVAPEE666 pKa = 3.64IGFEE670 pKa = 3.95IKK672 pKa = 10.76SPINFILKK680 pKa = 8.86TEE682 pKa = 4.0IQRR685 pKa = 11.84LHH687 pKa = 6.74GNLEE691 pKa = 3.99MQMEE695 pKa = 4.43KK696 pKa = 10.42PLRR699 pKa = 11.84EE700 pKa = 4.08AGIKK704 pKa = 9.87CC705 pKa = 3.76
Molecular weight: 81.44 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q6Q2W5|Q6Q2W5_9VIRU RNA-dependent RNA polymerase OS=Thielaviopsis basicola mitovirus OX=274584 PE=4 SV=1
MM1 pKa = 7.44IKK3 pKa = 10.43YY4 pKa = 7.0MKK6 pKa = 9.95SVRR9 pKa = 11.84LHH11 pKa = 4.55ITRR14 pKa = 11.84YY15 pKa = 8.93ICRR18 pKa = 11.84KK19 pKa = 8.83PLFINDD25 pKa = 3.55SGVSVDD31 pKa = 4.31KK32 pKa = 11.37SGFPTKK38 pKa = 10.53FIEE41 pKa = 5.17LKK43 pKa = 10.67KK44 pKa = 10.48FLNNHH49 pKa = 5.82EE50 pKa = 4.41SSICYY55 pKa = 7.68HH56 pKa = 5.15TRR58 pKa = 11.84LRR60 pKa = 11.84IILTLLLLPRR70 pKa = 11.84GLKK73 pKa = 7.37PTKK76 pKa = 10.27IEE78 pKa = 4.01DD79 pKa = 3.55AKK81 pKa = 10.72IKK83 pKa = 10.58PNFSSITLPFSGTRR97 pKa = 11.84MNPIPTWFIKK107 pKa = 10.62EE108 pKa = 3.88FVKK111 pKa = 10.65EE112 pKa = 3.97YY113 pKa = 10.64NLKK116 pKa = 10.58LEE118 pKa = 4.4KK119 pKa = 9.22PTYY122 pKa = 10.12DD123 pKa = 3.2LSKK126 pKa = 10.58HH127 pKa = 4.92YY128 pKa = 10.12LSPKK132 pKa = 8.55GGPMGKK138 pKa = 8.08STWSSIWSMYY148 pKa = 9.59ALKK151 pKa = 10.72HH152 pKa = 5.42EE153 pKa = 4.41LHH155 pKa = 6.61LSLSYY160 pKa = 10.78FLGEE164 pKa = 3.9AYY166 pKa = 7.57KK167 pKa = 10.11TLFYY171 pKa = 11.01LPYY174 pKa = 9.15LKK176 pKa = 10.86NMALSYY182 pKa = 10.94GHH184 pKa = 7.23DD185 pKa = 3.9VPPKK189 pKa = 8.34QWPSGKK195 pKa = 10.26LGIVKK200 pKa = 10.26DD201 pKa = 4.0PEE203 pKa = 4.01GKK205 pKa = 9.82RR206 pKa = 11.84RR207 pKa = 11.84IIAMVDD213 pKa = 3.06YY214 pKa = 10.6HH215 pKa = 6.58SQLVLRR221 pKa = 11.84SIHH224 pKa = 7.51DD225 pKa = 3.56GLLNKK230 pKa = 9.71LRR232 pKa = 11.84NLPQDD237 pKa = 2.95RR238 pKa = 11.84TYY240 pKa = 11.55NQDD243 pKa = 3.38PNNAWEE249 pKa = 4.04EE250 pKa = 4.07NKK252 pKa = 10.54EE253 pKa = 4.24CFHH256 pKa = 8.14SLDD259 pKa = 4.13LSSATDD265 pKa = 3.76RR266 pKa = 11.84FPVKK270 pKa = 10.18LQSRR274 pKa = 11.84LLTEE278 pKa = 4.7MYY280 pKa = 10.32SDD282 pKa = 3.98PSFGEE287 pKa = 3.26NWMNLLLNRR296 pKa = 11.84DD297 pKa = 3.63YY298 pKa = 11.49LLPEE302 pKa = 4.46EE303 pKa = 4.59GLSGEE308 pKa = 4.16RR309 pKa = 11.84LRR311 pKa = 11.84YY312 pKa = 9.52AVGQPMGAYY321 pKa = 9.89SSWAAFTLSHH331 pKa = 6.52HH332 pKa = 6.89LVVAWCTYY340 pKa = 9.64KK341 pKa = 10.44SKK343 pKa = 11.02KK344 pKa = 9.8VIRR347 pKa = 11.84SSQYY351 pKa = 10.25IILGDD356 pKa = 4.4DD357 pKa = 3.66IVIKK361 pKa = 11.03DD362 pKa = 3.33NDD364 pKa = 3.4IARR367 pKa = 11.84KK368 pKa = 10.13YY369 pKa = 9.03IGQMSKK375 pKa = 10.84LGVAISMQKK384 pKa = 7.76THH386 pKa = 5.51VSKK389 pKa = 10.08DD390 pKa = 3.06TYY392 pKa = 10.88EE393 pKa = 3.94FAKK396 pKa = 10.15RR397 pKa = 11.84WMHH400 pKa = 6.36KK401 pKa = 9.87GVEE404 pKa = 4.09ISGLPLKK411 pKa = 10.87GIFYY415 pKa = 10.02NISHH419 pKa = 6.48MRR421 pKa = 11.84RR422 pKa = 11.84MYY424 pKa = 10.28TIIFDD429 pKa = 3.78YY430 pKa = 10.47LQRR433 pKa = 11.84IPSKK437 pKa = 11.32SNLTSLQLFASCLSGIKK454 pKa = 9.8FGKK457 pKa = 9.6RR458 pKa = 11.84VYY460 pKa = 10.88SKK462 pKa = 11.56ALILKK467 pKa = 9.19LLRR470 pKa = 11.84NFNISLRR477 pKa = 11.84YY478 pKa = 9.65SYY480 pKa = 11.56GLLTPYY486 pKa = 10.41EE487 pKa = 4.09LRR489 pKa = 11.84EE490 pKa = 4.06YY491 pKa = 10.56ILTGFNMGDD500 pKa = 3.19EE501 pKa = 4.58VLPSDD506 pKa = 4.63NRR508 pKa = 11.84ILEE511 pKa = 4.13WFKK514 pKa = 11.4GIICDD519 pKa = 3.86GVAGKK524 pKa = 10.74VSTLSRR530 pKa = 11.84EE531 pKa = 3.89YY532 pKa = 9.73TKK534 pKa = 10.64EE535 pKa = 3.99VNSLEE540 pKa = 4.16EE541 pKa = 4.22HH542 pKa = 6.85RR543 pKa = 11.84SSQHH547 pKa = 5.33ILNSINIEE555 pKa = 3.63ITYY558 pKa = 10.73EE559 pKa = 3.89NLQWSPLIIGMFNHH573 pKa = 6.38INKK576 pKa = 9.19VSEE579 pKa = 4.31KK580 pKa = 10.23CGEE583 pKa = 3.88WLYY586 pKa = 11.67GDD588 pKa = 4.34VPLMDD593 pKa = 5.88IIQQFTLPSADD604 pKa = 3.27SLSRR608 pKa = 11.84KK609 pKa = 9.92DD610 pKa = 3.48RR611 pKa = 11.84DD612 pKa = 3.35INKK615 pKa = 10.27KK616 pKa = 9.59IDD618 pKa = 3.71EE619 pKa = 4.67LDD621 pKa = 3.44SLIFKK626 pKa = 9.95SIKK629 pKa = 9.9RR630 pKa = 11.84HH631 pKa = 6.22FSDD634 pKa = 4.3QPYY637 pKa = 9.32PDD639 pKa = 3.92HH640 pKa = 7.65FYY642 pKa = 10.09GTRR645 pKa = 11.84SNGDD649 pKa = 3.06KK650 pKa = 10.77NLYY653 pKa = 9.05PGLGALDD660 pKa = 3.61VNVAPEE666 pKa = 3.64IGFEE670 pKa = 3.95IKK672 pKa = 10.76SPINFILKK680 pKa = 8.86TEE682 pKa = 4.0IQRR685 pKa = 11.84LHH687 pKa = 6.74GNLEE691 pKa = 3.99MQMEE695 pKa = 4.43KK696 pKa = 10.42PLRR699 pKa = 11.84EE700 pKa = 4.08AGIKK704 pKa = 9.87CC705 pKa = 3.76
MM1 pKa = 7.44IKK3 pKa = 10.43YY4 pKa = 7.0MKK6 pKa = 9.95SVRR9 pKa = 11.84LHH11 pKa = 4.55ITRR14 pKa = 11.84YY15 pKa = 8.93ICRR18 pKa = 11.84KK19 pKa = 8.83PLFINDD25 pKa = 3.55SGVSVDD31 pKa = 4.31KK32 pKa = 11.37SGFPTKK38 pKa = 10.53FIEE41 pKa = 5.17LKK43 pKa = 10.67KK44 pKa = 10.48FLNNHH49 pKa = 5.82EE50 pKa = 4.41SSICYY55 pKa = 7.68HH56 pKa = 5.15TRR58 pKa = 11.84LRR60 pKa = 11.84IILTLLLLPRR70 pKa = 11.84GLKK73 pKa = 7.37PTKK76 pKa = 10.27IEE78 pKa = 4.01DD79 pKa = 3.55AKK81 pKa = 10.72IKK83 pKa = 10.58PNFSSITLPFSGTRR97 pKa = 11.84MNPIPTWFIKK107 pKa = 10.62EE108 pKa = 3.88FVKK111 pKa = 10.65EE112 pKa = 3.97YY113 pKa = 10.64NLKK116 pKa = 10.58LEE118 pKa = 4.4KK119 pKa = 9.22PTYY122 pKa = 10.12DD123 pKa = 3.2LSKK126 pKa = 10.58HH127 pKa = 4.92YY128 pKa = 10.12LSPKK132 pKa = 8.55GGPMGKK138 pKa = 8.08STWSSIWSMYY148 pKa = 9.59ALKK151 pKa = 10.72HH152 pKa = 5.42EE153 pKa = 4.41LHH155 pKa = 6.61LSLSYY160 pKa = 10.78FLGEE164 pKa = 3.9AYY166 pKa = 7.57KK167 pKa = 10.11TLFYY171 pKa = 11.01LPYY174 pKa = 9.15LKK176 pKa = 10.86NMALSYY182 pKa = 10.94GHH184 pKa = 7.23DD185 pKa = 3.9VPPKK189 pKa = 8.34QWPSGKK195 pKa = 10.26LGIVKK200 pKa = 10.26DD201 pKa = 4.0PEE203 pKa = 4.01GKK205 pKa = 9.82RR206 pKa = 11.84RR207 pKa = 11.84IIAMVDD213 pKa = 3.06YY214 pKa = 10.6HH215 pKa = 6.58SQLVLRR221 pKa = 11.84SIHH224 pKa = 7.51DD225 pKa = 3.56GLLNKK230 pKa = 9.71LRR232 pKa = 11.84NLPQDD237 pKa = 2.95RR238 pKa = 11.84TYY240 pKa = 11.55NQDD243 pKa = 3.38PNNAWEE249 pKa = 4.04EE250 pKa = 4.07NKK252 pKa = 10.54EE253 pKa = 4.24CFHH256 pKa = 8.14SLDD259 pKa = 4.13LSSATDD265 pKa = 3.76RR266 pKa = 11.84FPVKK270 pKa = 10.18LQSRR274 pKa = 11.84LLTEE278 pKa = 4.7MYY280 pKa = 10.32SDD282 pKa = 3.98PSFGEE287 pKa = 3.26NWMNLLLNRR296 pKa = 11.84DD297 pKa = 3.63YY298 pKa = 11.49LLPEE302 pKa = 4.46EE303 pKa = 4.59GLSGEE308 pKa = 4.16RR309 pKa = 11.84LRR311 pKa = 11.84YY312 pKa = 9.52AVGQPMGAYY321 pKa = 9.89SSWAAFTLSHH331 pKa = 6.52HH332 pKa = 6.89LVVAWCTYY340 pKa = 9.64KK341 pKa = 10.44SKK343 pKa = 11.02KK344 pKa = 9.8VIRR347 pKa = 11.84SSQYY351 pKa = 10.25IILGDD356 pKa = 4.4DD357 pKa = 3.66IVIKK361 pKa = 11.03DD362 pKa = 3.33NDD364 pKa = 3.4IARR367 pKa = 11.84KK368 pKa = 10.13YY369 pKa = 9.03IGQMSKK375 pKa = 10.84LGVAISMQKK384 pKa = 7.76THH386 pKa = 5.51VSKK389 pKa = 10.08DD390 pKa = 3.06TYY392 pKa = 10.88EE393 pKa = 3.94FAKK396 pKa = 10.15RR397 pKa = 11.84WMHH400 pKa = 6.36KK401 pKa = 9.87GVEE404 pKa = 4.09ISGLPLKK411 pKa = 10.87GIFYY415 pKa = 10.02NISHH419 pKa = 6.48MRR421 pKa = 11.84RR422 pKa = 11.84MYY424 pKa = 10.28TIIFDD429 pKa = 3.78YY430 pKa = 10.47LQRR433 pKa = 11.84IPSKK437 pKa = 11.32SNLTSLQLFASCLSGIKK454 pKa = 9.8FGKK457 pKa = 9.6RR458 pKa = 11.84VYY460 pKa = 10.88SKK462 pKa = 11.56ALILKK467 pKa = 9.19LLRR470 pKa = 11.84NFNISLRR477 pKa = 11.84YY478 pKa = 9.65SYY480 pKa = 11.56GLLTPYY486 pKa = 10.41EE487 pKa = 4.09LRR489 pKa = 11.84EE490 pKa = 4.06YY491 pKa = 10.56ILTGFNMGDD500 pKa = 3.19EE501 pKa = 4.58VLPSDD506 pKa = 4.63NRR508 pKa = 11.84ILEE511 pKa = 4.13WFKK514 pKa = 11.4GIICDD519 pKa = 3.86GVAGKK524 pKa = 10.74VSTLSRR530 pKa = 11.84EE531 pKa = 3.89YY532 pKa = 9.73TKK534 pKa = 10.64EE535 pKa = 3.99VNSLEE540 pKa = 4.16EE541 pKa = 4.22HH542 pKa = 6.85RR543 pKa = 11.84SSQHH547 pKa = 5.33ILNSINIEE555 pKa = 3.63ITYY558 pKa = 10.73EE559 pKa = 3.89NLQWSPLIIGMFNHH573 pKa = 6.38INKK576 pKa = 9.19VSEE579 pKa = 4.31KK580 pKa = 10.23CGEE583 pKa = 3.88WLYY586 pKa = 11.67GDD588 pKa = 4.34VPLMDD593 pKa = 5.88IIQQFTLPSADD604 pKa = 3.27SLSRR608 pKa = 11.84KK609 pKa = 9.92DD610 pKa = 3.48RR611 pKa = 11.84DD612 pKa = 3.35INKK615 pKa = 10.27KK616 pKa = 9.59IDD618 pKa = 3.71EE619 pKa = 4.67LDD621 pKa = 3.44SLIFKK626 pKa = 9.95SIKK629 pKa = 9.9RR630 pKa = 11.84HH631 pKa = 6.22FSDD634 pKa = 4.3QPYY637 pKa = 9.32PDD639 pKa = 3.92HH640 pKa = 7.65FYY642 pKa = 10.09GTRR645 pKa = 11.84SNGDD649 pKa = 3.06KK650 pKa = 10.77NLYY653 pKa = 9.05PGLGALDD660 pKa = 3.61VNVAPEE666 pKa = 3.64IGFEE670 pKa = 3.95IKK672 pKa = 10.76SPINFILKK680 pKa = 8.86TEE682 pKa = 4.0IQRR685 pKa = 11.84LHH687 pKa = 6.74GNLEE691 pKa = 3.99MQMEE695 pKa = 4.43KK696 pKa = 10.42PLRR699 pKa = 11.84EE700 pKa = 4.08AGIKK704 pKa = 9.87CC705 pKa = 3.76
Molecular weight: 81.44 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
705 |
705 |
705 |
705.0 |
81.44 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
3.121 ± 0.0 | 1.135 ± 0.0 |
4.823 ± 0.0 | 5.39 ± 0.0 |
4.113 ± 0.0 | 5.957 ± 0.0 |
2.979 ± 0.0 | 8.227 ± 0.0 |
8.369 ± 0.0 | 11.915 ± 0.0 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.837 ± 0.0 | 4.965 ± 0.0 |
4.965 ± 0.0 | 2.553 ± 0.0 |
5.106 ± 0.0 | 9.078 ± 0.0 |
3.972 ± 0.0 | 3.688 ± 0.0 |
1.702 ± 0.0 | 5.106 ± 0.0 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
