
Beihai narna-like virus 6
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.29
Get precalculated fractions of proteins
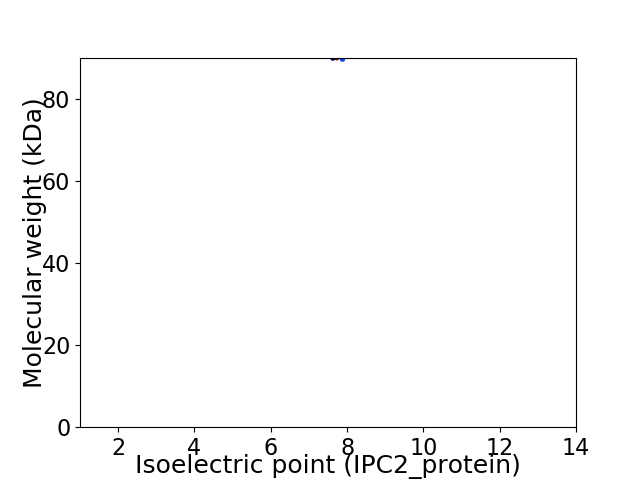
Virtual 2D-PAGE plot for 1 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KI33|A0A1L3KI33_9VIRU RNA-dependent RNA polymerase OS=Beihai narna-like virus 6 OX=1922458 PE=4 SV=1
MM1 pKa = 7.5FCRR4 pKa = 11.84SVWDD8 pKa = 3.84VLCDD12 pKa = 3.46ADD14 pKa = 4.27QVFRR18 pKa = 11.84YY19 pKa = 8.85MPARR23 pKa = 11.84CFLDD27 pKa = 3.94GFLPLLMRR35 pKa = 11.84AKK37 pKa = 10.74DD38 pKa = 3.38EE39 pKa = 4.35AEE41 pKa = 3.77QVKK44 pKa = 7.72WQKK47 pKa = 11.29YY48 pKa = 5.12LTCWPMAKK56 pKa = 10.15FLRR59 pKa = 11.84QTSMPEE65 pKa = 3.73VPAGLEE71 pKa = 3.89EE72 pKa = 5.28AIGSSFHH79 pKa = 7.15FPLTGAARR87 pKa = 11.84RR88 pKa = 11.84HH89 pKa = 5.45LRR91 pKa = 11.84NLLASRR97 pKa = 11.84DD98 pKa = 3.74SEE100 pKa = 3.89IRR102 pKa = 11.84PTQVCWAILQGVKK115 pKa = 10.22RR116 pKa = 11.84GCAEE120 pKa = 3.69VSEE123 pKa = 4.43DD124 pKa = 4.46FIYY127 pKa = 9.48ATMLKK132 pKa = 9.83HH133 pKa = 6.44KK134 pKa = 10.7ASLTQTLPEE143 pKa = 3.99MSEE146 pKa = 3.95EE147 pKa = 3.84ATEE150 pKa = 4.1LVRR153 pKa = 11.84QKK155 pKa = 10.32FRR157 pKa = 11.84NIWRR161 pKa = 11.84PSFRR165 pKa = 11.84RR166 pKa = 11.84RR167 pKa = 11.84QWTGYY172 pKa = 7.29GTSRR176 pKa = 11.84QMEE179 pKa = 4.37RR180 pKa = 11.84DD181 pKa = 3.58CSISGNPNFNACYY194 pKa = 9.78EE195 pKa = 4.35VTRR198 pKa = 11.84SQGGRR203 pKa = 11.84CRR205 pKa = 11.84AVRR208 pKa = 11.84STIIRR213 pKa = 11.84YY214 pKa = 8.65LRR216 pKa = 11.84EE217 pKa = 3.78EE218 pKa = 3.78WGMDD222 pKa = 2.97SDD224 pKa = 4.56NRR226 pKa = 11.84LLYY229 pKa = 10.8RR230 pKa = 11.84MTEE233 pKa = 3.98VAPGEE238 pKa = 4.38VVSEE242 pKa = 4.21YY243 pKa = 10.23IGPEE247 pKa = 3.75FLPEE251 pKa = 4.2VSQKK255 pKa = 10.62LAEE258 pKa = 4.02RR259 pKa = 11.84WALKK263 pKa = 10.44DD264 pKa = 3.54LEE266 pKa = 4.5RR267 pKa = 11.84LGGTCRR273 pKa = 11.84ATVCAILEE281 pKa = 4.05PLKK284 pKa = 10.95CRR286 pKa = 11.84LITKK290 pKa = 10.02GSALPYY296 pKa = 9.82FAAQALQKK304 pKa = 11.23AMWQRR309 pKa = 11.84LQAFPCFKK317 pKa = 10.41LTGCPLDD324 pKa = 4.27ASMLQGLLDD333 pKa = 3.5QEE335 pKa = 4.39EE336 pKa = 4.28RR337 pKa = 11.84LGLDD341 pKa = 3.14FTHH344 pKa = 6.99WVSGDD349 pKa = 3.32YY350 pKa = 10.73SAATDD355 pKa = 3.92GLSQQVNRR363 pKa = 11.84LCLEE367 pKa = 3.9EE368 pKa = 4.7ALRR371 pKa = 11.84GCNASEE377 pKa = 3.81ALKK380 pKa = 10.5IIARR384 pKa = 11.84AVLGNHH390 pKa = 6.64RR391 pKa = 11.84IEE393 pKa = 4.44YY394 pKa = 6.88PTEE397 pKa = 3.9FEE399 pKa = 4.25IDD401 pKa = 3.79GMMQQNGQLMGSPLSFPVLCSINVAAYY428 pKa = 7.28WCALEE433 pKa = 5.16EE434 pKa = 4.0YY435 pKa = 9.38TGRR438 pKa = 11.84KK439 pKa = 8.19FQLEE443 pKa = 4.24DD444 pKa = 3.98LPCLVNGDD452 pKa = 4.86DD453 pKa = 3.24ICFRR457 pKa = 11.84ANPEE461 pKa = 4.4FYY463 pKa = 10.7GIWQRR468 pKa = 11.84WVRR471 pKa = 11.84EE472 pKa = 3.95VGFTLSPGKK481 pKa = 10.4NYY483 pKa = 9.68IHH485 pKa = 6.16EE486 pKa = 4.53HH487 pKa = 5.02FVTVNSEE494 pKa = 4.46GYY496 pKa = 9.0VHH498 pKa = 6.36QKK500 pKa = 9.18HH501 pKa = 5.95GKK503 pKa = 9.21VPFHH507 pKa = 6.71KK508 pKa = 10.96VNFLNTGLLYY518 pKa = 10.57SGKK521 pKa = 9.92SVEE524 pKa = 5.53RR525 pKa = 11.84KK526 pKa = 9.35IDD528 pKa = 3.31WGDD531 pKa = 3.34QPAIKK536 pKa = 10.47VGLRR540 pKa = 11.84PEE542 pKa = 3.97NRR544 pKa = 11.84EE545 pKa = 3.62MPFTQKK551 pKa = 9.85VNRR554 pKa = 11.84VIEE557 pKa = 4.48EE558 pKa = 4.22SCSPTRR564 pKa = 11.84TLLRR568 pKa = 11.84VHH570 pKa = 7.29RR571 pKa = 11.84YY572 pKa = 8.43FRR574 pKa = 11.84DD575 pKa = 3.47EE576 pKa = 4.1IKK578 pKa = 10.77YY579 pKa = 8.06HH580 pKa = 5.13TLNGEE585 pKa = 4.42INMHH589 pKa = 6.9AAPEE593 pKa = 4.15LGGLGIVLPEE603 pKa = 4.37GSDD606 pKa = 3.31TRR608 pKa = 11.84FTAWQCRR615 pKa = 11.84AAGYY619 pKa = 9.02LASRR623 pKa = 11.84WKK625 pKa = 10.44SLEE628 pKa = 3.82FGSVVEE634 pKa = 5.23GIPGPLTQEE643 pKa = 4.08EE644 pKa = 4.34AKK646 pKa = 10.71HH647 pKa = 6.44RR648 pKa = 11.84IVDD651 pKa = 4.05LNRR654 pKa = 11.84PLGVEE659 pKa = 3.95GRR661 pKa = 11.84VTYY664 pKa = 7.93QQKK667 pKa = 10.55KK668 pKa = 8.98SAALTWQAPVKK679 pKa = 9.44PGRR682 pKa = 11.84VVVRR686 pKa = 11.84EE687 pKa = 3.76KK688 pKa = 11.22LEE690 pKa = 3.79PLRR693 pKa = 11.84EE694 pKa = 4.16GEE696 pKa = 4.12EE697 pKa = 4.12RR698 pKa = 11.84IEE700 pKa = 4.42PDD702 pKa = 2.96SSPLRR707 pKa = 11.84NYY709 pKa = 9.9QGPPTSANGEE719 pKa = 4.17WKK721 pKa = 10.35IKK723 pKa = 10.77GLDD726 pKa = 3.61SEE728 pKa = 4.57CRR730 pKa = 11.84QAIRR734 pKa = 11.84EE735 pKa = 4.02YY736 pKa = 10.47RR737 pKa = 11.84GSGVNRR743 pKa = 11.84PLRR746 pKa = 11.84WSRR749 pKa = 11.84EE750 pKa = 3.84VRR752 pKa = 11.84SAPSSILDD760 pKa = 3.3RR761 pKa = 11.84LVRR764 pKa = 11.84SDD766 pKa = 3.47STSVLNTWIGAEE778 pKa = 4.18GQVHH782 pKa = 6.28SFLSKK787 pKa = 10.23PVLVGG792 pKa = 3.35
MM1 pKa = 7.5FCRR4 pKa = 11.84SVWDD8 pKa = 3.84VLCDD12 pKa = 3.46ADD14 pKa = 4.27QVFRR18 pKa = 11.84YY19 pKa = 8.85MPARR23 pKa = 11.84CFLDD27 pKa = 3.94GFLPLLMRR35 pKa = 11.84AKK37 pKa = 10.74DD38 pKa = 3.38EE39 pKa = 4.35AEE41 pKa = 3.77QVKK44 pKa = 7.72WQKK47 pKa = 11.29YY48 pKa = 5.12LTCWPMAKK56 pKa = 10.15FLRR59 pKa = 11.84QTSMPEE65 pKa = 3.73VPAGLEE71 pKa = 3.89EE72 pKa = 5.28AIGSSFHH79 pKa = 7.15FPLTGAARR87 pKa = 11.84RR88 pKa = 11.84HH89 pKa = 5.45LRR91 pKa = 11.84NLLASRR97 pKa = 11.84DD98 pKa = 3.74SEE100 pKa = 3.89IRR102 pKa = 11.84PTQVCWAILQGVKK115 pKa = 10.22RR116 pKa = 11.84GCAEE120 pKa = 3.69VSEE123 pKa = 4.43DD124 pKa = 4.46FIYY127 pKa = 9.48ATMLKK132 pKa = 9.83HH133 pKa = 6.44KK134 pKa = 10.7ASLTQTLPEE143 pKa = 3.99MSEE146 pKa = 3.95EE147 pKa = 3.84ATEE150 pKa = 4.1LVRR153 pKa = 11.84QKK155 pKa = 10.32FRR157 pKa = 11.84NIWRR161 pKa = 11.84PSFRR165 pKa = 11.84RR166 pKa = 11.84RR167 pKa = 11.84QWTGYY172 pKa = 7.29GTSRR176 pKa = 11.84QMEE179 pKa = 4.37RR180 pKa = 11.84DD181 pKa = 3.58CSISGNPNFNACYY194 pKa = 9.78EE195 pKa = 4.35VTRR198 pKa = 11.84SQGGRR203 pKa = 11.84CRR205 pKa = 11.84AVRR208 pKa = 11.84STIIRR213 pKa = 11.84YY214 pKa = 8.65LRR216 pKa = 11.84EE217 pKa = 3.78EE218 pKa = 3.78WGMDD222 pKa = 2.97SDD224 pKa = 4.56NRR226 pKa = 11.84LLYY229 pKa = 10.8RR230 pKa = 11.84MTEE233 pKa = 3.98VAPGEE238 pKa = 4.38VVSEE242 pKa = 4.21YY243 pKa = 10.23IGPEE247 pKa = 3.75FLPEE251 pKa = 4.2VSQKK255 pKa = 10.62LAEE258 pKa = 4.02RR259 pKa = 11.84WALKK263 pKa = 10.44DD264 pKa = 3.54LEE266 pKa = 4.5RR267 pKa = 11.84LGGTCRR273 pKa = 11.84ATVCAILEE281 pKa = 4.05PLKK284 pKa = 10.95CRR286 pKa = 11.84LITKK290 pKa = 10.02GSALPYY296 pKa = 9.82FAAQALQKK304 pKa = 11.23AMWQRR309 pKa = 11.84LQAFPCFKK317 pKa = 10.41LTGCPLDD324 pKa = 4.27ASMLQGLLDD333 pKa = 3.5QEE335 pKa = 4.39EE336 pKa = 4.28RR337 pKa = 11.84LGLDD341 pKa = 3.14FTHH344 pKa = 6.99WVSGDD349 pKa = 3.32YY350 pKa = 10.73SAATDD355 pKa = 3.92GLSQQVNRR363 pKa = 11.84LCLEE367 pKa = 3.9EE368 pKa = 4.7ALRR371 pKa = 11.84GCNASEE377 pKa = 3.81ALKK380 pKa = 10.5IIARR384 pKa = 11.84AVLGNHH390 pKa = 6.64RR391 pKa = 11.84IEE393 pKa = 4.44YY394 pKa = 6.88PTEE397 pKa = 3.9FEE399 pKa = 4.25IDD401 pKa = 3.79GMMQQNGQLMGSPLSFPVLCSINVAAYY428 pKa = 7.28WCALEE433 pKa = 5.16EE434 pKa = 4.0YY435 pKa = 9.38TGRR438 pKa = 11.84KK439 pKa = 8.19FQLEE443 pKa = 4.24DD444 pKa = 3.98LPCLVNGDD452 pKa = 4.86DD453 pKa = 3.24ICFRR457 pKa = 11.84ANPEE461 pKa = 4.4FYY463 pKa = 10.7GIWQRR468 pKa = 11.84WVRR471 pKa = 11.84EE472 pKa = 3.95VGFTLSPGKK481 pKa = 10.4NYY483 pKa = 9.68IHH485 pKa = 6.16EE486 pKa = 4.53HH487 pKa = 5.02FVTVNSEE494 pKa = 4.46GYY496 pKa = 9.0VHH498 pKa = 6.36QKK500 pKa = 9.18HH501 pKa = 5.95GKK503 pKa = 9.21VPFHH507 pKa = 6.71KK508 pKa = 10.96VNFLNTGLLYY518 pKa = 10.57SGKK521 pKa = 9.92SVEE524 pKa = 5.53RR525 pKa = 11.84KK526 pKa = 9.35IDD528 pKa = 3.31WGDD531 pKa = 3.34QPAIKK536 pKa = 10.47VGLRR540 pKa = 11.84PEE542 pKa = 3.97NRR544 pKa = 11.84EE545 pKa = 3.62MPFTQKK551 pKa = 9.85VNRR554 pKa = 11.84VIEE557 pKa = 4.48EE558 pKa = 4.22SCSPTRR564 pKa = 11.84TLLRR568 pKa = 11.84VHH570 pKa = 7.29RR571 pKa = 11.84YY572 pKa = 8.43FRR574 pKa = 11.84DD575 pKa = 3.47EE576 pKa = 4.1IKK578 pKa = 10.77YY579 pKa = 8.06HH580 pKa = 5.13TLNGEE585 pKa = 4.42INMHH589 pKa = 6.9AAPEE593 pKa = 4.15LGGLGIVLPEE603 pKa = 4.37GSDD606 pKa = 3.31TRR608 pKa = 11.84FTAWQCRR615 pKa = 11.84AAGYY619 pKa = 9.02LASRR623 pKa = 11.84WKK625 pKa = 10.44SLEE628 pKa = 3.82FGSVVEE634 pKa = 5.23GIPGPLTQEE643 pKa = 4.08EE644 pKa = 4.34AKK646 pKa = 10.71HH647 pKa = 6.44RR648 pKa = 11.84IVDD651 pKa = 4.05LNRR654 pKa = 11.84PLGVEE659 pKa = 3.95GRR661 pKa = 11.84VTYY664 pKa = 7.93QQKK667 pKa = 10.55KK668 pKa = 8.98SAALTWQAPVKK679 pKa = 9.44PGRR682 pKa = 11.84VVVRR686 pKa = 11.84EE687 pKa = 3.76KK688 pKa = 11.22LEE690 pKa = 3.79PLRR693 pKa = 11.84EE694 pKa = 4.16GEE696 pKa = 4.12EE697 pKa = 4.12RR698 pKa = 11.84IEE700 pKa = 4.42PDD702 pKa = 2.96SSPLRR707 pKa = 11.84NYY709 pKa = 9.9QGPPTSANGEE719 pKa = 4.17WKK721 pKa = 10.35IKK723 pKa = 10.77GLDD726 pKa = 3.61SEE728 pKa = 4.57CRR730 pKa = 11.84QAIRR734 pKa = 11.84EE735 pKa = 4.02YY736 pKa = 10.47RR737 pKa = 11.84GSGVNRR743 pKa = 11.84PLRR746 pKa = 11.84WSRR749 pKa = 11.84EE750 pKa = 3.84VRR752 pKa = 11.84SAPSSILDD760 pKa = 3.3RR761 pKa = 11.84LVRR764 pKa = 11.84SDD766 pKa = 3.47STSVLNTWIGAEE778 pKa = 4.18GQVHH782 pKa = 6.28SFLSKK787 pKa = 10.23PVLVGG792 pKa = 3.35
Molecular weight: 90.01 kDa
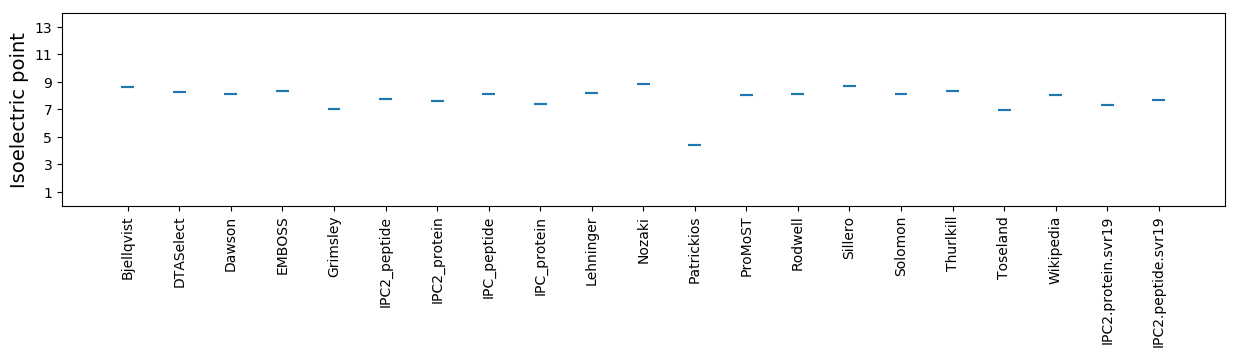
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KI33|A0A1L3KI33_9VIRU RNA-dependent RNA polymerase OS=Beihai narna-like virus 6 OX=1922458 PE=4 SV=1
MM1 pKa = 7.5FCRR4 pKa = 11.84SVWDD8 pKa = 3.84VLCDD12 pKa = 3.46ADD14 pKa = 4.27QVFRR18 pKa = 11.84YY19 pKa = 8.85MPARR23 pKa = 11.84CFLDD27 pKa = 3.94GFLPLLMRR35 pKa = 11.84AKK37 pKa = 10.74DD38 pKa = 3.38EE39 pKa = 4.35AEE41 pKa = 3.77QVKK44 pKa = 7.72WQKK47 pKa = 11.29YY48 pKa = 5.12LTCWPMAKK56 pKa = 10.15FLRR59 pKa = 11.84QTSMPEE65 pKa = 3.73VPAGLEE71 pKa = 3.89EE72 pKa = 5.28AIGSSFHH79 pKa = 7.15FPLTGAARR87 pKa = 11.84RR88 pKa = 11.84HH89 pKa = 5.45LRR91 pKa = 11.84NLLASRR97 pKa = 11.84DD98 pKa = 3.74SEE100 pKa = 3.89IRR102 pKa = 11.84PTQVCWAILQGVKK115 pKa = 10.22RR116 pKa = 11.84GCAEE120 pKa = 3.69VSEE123 pKa = 4.43DD124 pKa = 4.46FIYY127 pKa = 9.48ATMLKK132 pKa = 9.83HH133 pKa = 6.44KK134 pKa = 10.7ASLTQTLPEE143 pKa = 3.99MSEE146 pKa = 3.95EE147 pKa = 3.84ATEE150 pKa = 4.1LVRR153 pKa = 11.84QKK155 pKa = 10.32FRR157 pKa = 11.84NIWRR161 pKa = 11.84PSFRR165 pKa = 11.84RR166 pKa = 11.84RR167 pKa = 11.84QWTGYY172 pKa = 7.29GTSRR176 pKa = 11.84QMEE179 pKa = 4.37RR180 pKa = 11.84DD181 pKa = 3.58CSISGNPNFNACYY194 pKa = 9.78EE195 pKa = 4.35VTRR198 pKa = 11.84SQGGRR203 pKa = 11.84CRR205 pKa = 11.84AVRR208 pKa = 11.84STIIRR213 pKa = 11.84YY214 pKa = 8.65LRR216 pKa = 11.84EE217 pKa = 3.78EE218 pKa = 3.78WGMDD222 pKa = 2.97SDD224 pKa = 4.56NRR226 pKa = 11.84LLYY229 pKa = 10.8RR230 pKa = 11.84MTEE233 pKa = 3.98VAPGEE238 pKa = 4.38VVSEE242 pKa = 4.21YY243 pKa = 10.23IGPEE247 pKa = 3.75FLPEE251 pKa = 4.2VSQKK255 pKa = 10.62LAEE258 pKa = 4.02RR259 pKa = 11.84WALKK263 pKa = 10.44DD264 pKa = 3.54LEE266 pKa = 4.5RR267 pKa = 11.84LGGTCRR273 pKa = 11.84ATVCAILEE281 pKa = 4.05PLKK284 pKa = 10.95CRR286 pKa = 11.84LITKK290 pKa = 10.02GSALPYY296 pKa = 9.82FAAQALQKK304 pKa = 11.23AMWQRR309 pKa = 11.84LQAFPCFKK317 pKa = 10.41LTGCPLDD324 pKa = 4.27ASMLQGLLDD333 pKa = 3.5QEE335 pKa = 4.39EE336 pKa = 4.28RR337 pKa = 11.84LGLDD341 pKa = 3.14FTHH344 pKa = 6.99WVSGDD349 pKa = 3.32YY350 pKa = 10.73SAATDD355 pKa = 3.92GLSQQVNRR363 pKa = 11.84LCLEE367 pKa = 3.9EE368 pKa = 4.7ALRR371 pKa = 11.84GCNASEE377 pKa = 3.81ALKK380 pKa = 10.5IIARR384 pKa = 11.84AVLGNHH390 pKa = 6.64RR391 pKa = 11.84IEE393 pKa = 4.44YY394 pKa = 6.88PTEE397 pKa = 3.9FEE399 pKa = 4.25IDD401 pKa = 3.79GMMQQNGQLMGSPLSFPVLCSINVAAYY428 pKa = 7.28WCALEE433 pKa = 5.16EE434 pKa = 4.0YY435 pKa = 9.38TGRR438 pKa = 11.84KK439 pKa = 8.19FQLEE443 pKa = 4.24DD444 pKa = 3.98LPCLVNGDD452 pKa = 4.86DD453 pKa = 3.24ICFRR457 pKa = 11.84ANPEE461 pKa = 4.4FYY463 pKa = 10.7GIWQRR468 pKa = 11.84WVRR471 pKa = 11.84EE472 pKa = 3.95VGFTLSPGKK481 pKa = 10.4NYY483 pKa = 9.68IHH485 pKa = 6.16EE486 pKa = 4.53HH487 pKa = 5.02FVTVNSEE494 pKa = 4.46GYY496 pKa = 9.0VHH498 pKa = 6.36QKK500 pKa = 9.18HH501 pKa = 5.95GKK503 pKa = 9.21VPFHH507 pKa = 6.71KK508 pKa = 10.96VNFLNTGLLYY518 pKa = 10.57SGKK521 pKa = 9.92SVEE524 pKa = 5.53RR525 pKa = 11.84KK526 pKa = 9.35IDD528 pKa = 3.31WGDD531 pKa = 3.34QPAIKK536 pKa = 10.47VGLRR540 pKa = 11.84PEE542 pKa = 3.97NRR544 pKa = 11.84EE545 pKa = 3.62MPFTQKK551 pKa = 9.85VNRR554 pKa = 11.84VIEE557 pKa = 4.48EE558 pKa = 4.22SCSPTRR564 pKa = 11.84TLLRR568 pKa = 11.84VHH570 pKa = 7.29RR571 pKa = 11.84YY572 pKa = 8.43FRR574 pKa = 11.84DD575 pKa = 3.47EE576 pKa = 4.1IKK578 pKa = 10.77YY579 pKa = 8.06HH580 pKa = 5.13TLNGEE585 pKa = 4.42INMHH589 pKa = 6.9AAPEE593 pKa = 4.15LGGLGIVLPEE603 pKa = 4.37GSDD606 pKa = 3.31TRR608 pKa = 11.84FTAWQCRR615 pKa = 11.84AAGYY619 pKa = 9.02LASRR623 pKa = 11.84WKK625 pKa = 10.44SLEE628 pKa = 3.82FGSVVEE634 pKa = 5.23GIPGPLTQEE643 pKa = 4.08EE644 pKa = 4.34AKK646 pKa = 10.71HH647 pKa = 6.44RR648 pKa = 11.84IVDD651 pKa = 4.05LNRR654 pKa = 11.84PLGVEE659 pKa = 3.95GRR661 pKa = 11.84VTYY664 pKa = 7.93QQKK667 pKa = 10.55KK668 pKa = 8.98SAALTWQAPVKK679 pKa = 9.44PGRR682 pKa = 11.84VVVRR686 pKa = 11.84EE687 pKa = 3.76KK688 pKa = 11.22LEE690 pKa = 3.79PLRR693 pKa = 11.84EE694 pKa = 4.16GEE696 pKa = 4.12EE697 pKa = 4.12RR698 pKa = 11.84IEE700 pKa = 4.42PDD702 pKa = 2.96SSPLRR707 pKa = 11.84NYY709 pKa = 9.9QGPPTSANGEE719 pKa = 4.17WKK721 pKa = 10.35IKK723 pKa = 10.77GLDD726 pKa = 3.61SEE728 pKa = 4.57CRR730 pKa = 11.84QAIRR734 pKa = 11.84EE735 pKa = 4.02YY736 pKa = 10.47RR737 pKa = 11.84GSGVNRR743 pKa = 11.84PLRR746 pKa = 11.84WSRR749 pKa = 11.84EE750 pKa = 3.84VRR752 pKa = 11.84SAPSSILDD760 pKa = 3.3RR761 pKa = 11.84LVRR764 pKa = 11.84SDD766 pKa = 3.47STSVLNTWIGAEE778 pKa = 4.18GQVHH782 pKa = 6.28SFLSKK787 pKa = 10.23PVLVGG792 pKa = 3.35
MM1 pKa = 7.5FCRR4 pKa = 11.84SVWDD8 pKa = 3.84VLCDD12 pKa = 3.46ADD14 pKa = 4.27QVFRR18 pKa = 11.84YY19 pKa = 8.85MPARR23 pKa = 11.84CFLDD27 pKa = 3.94GFLPLLMRR35 pKa = 11.84AKK37 pKa = 10.74DD38 pKa = 3.38EE39 pKa = 4.35AEE41 pKa = 3.77QVKK44 pKa = 7.72WQKK47 pKa = 11.29YY48 pKa = 5.12LTCWPMAKK56 pKa = 10.15FLRR59 pKa = 11.84QTSMPEE65 pKa = 3.73VPAGLEE71 pKa = 3.89EE72 pKa = 5.28AIGSSFHH79 pKa = 7.15FPLTGAARR87 pKa = 11.84RR88 pKa = 11.84HH89 pKa = 5.45LRR91 pKa = 11.84NLLASRR97 pKa = 11.84DD98 pKa = 3.74SEE100 pKa = 3.89IRR102 pKa = 11.84PTQVCWAILQGVKK115 pKa = 10.22RR116 pKa = 11.84GCAEE120 pKa = 3.69VSEE123 pKa = 4.43DD124 pKa = 4.46FIYY127 pKa = 9.48ATMLKK132 pKa = 9.83HH133 pKa = 6.44KK134 pKa = 10.7ASLTQTLPEE143 pKa = 3.99MSEE146 pKa = 3.95EE147 pKa = 3.84ATEE150 pKa = 4.1LVRR153 pKa = 11.84QKK155 pKa = 10.32FRR157 pKa = 11.84NIWRR161 pKa = 11.84PSFRR165 pKa = 11.84RR166 pKa = 11.84RR167 pKa = 11.84QWTGYY172 pKa = 7.29GTSRR176 pKa = 11.84QMEE179 pKa = 4.37RR180 pKa = 11.84DD181 pKa = 3.58CSISGNPNFNACYY194 pKa = 9.78EE195 pKa = 4.35VTRR198 pKa = 11.84SQGGRR203 pKa = 11.84CRR205 pKa = 11.84AVRR208 pKa = 11.84STIIRR213 pKa = 11.84YY214 pKa = 8.65LRR216 pKa = 11.84EE217 pKa = 3.78EE218 pKa = 3.78WGMDD222 pKa = 2.97SDD224 pKa = 4.56NRR226 pKa = 11.84LLYY229 pKa = 10.8RR230 pKa = 11.84MTEE233 pKa = 3.98VAPGEE238 pKa = 4.38VVSEE242 pKa = 4.21YY243 pKa = 10.23IGPEE247 pKa = 3.75FLPEE251 pKa = 4.2VSQKK255 pKa = 10.62LAEE258 pKa = 4.02RR259 pKa = 11.84WALKK263 pKa = 10.44DD264 pKa = 3.54LEE266 pKa = 4.5RR267 pKa = 11.84LGGTCRR273 pKa = 11.84ATVCAILEE281 pKa = 4.05PLKK284 pKa = 10.95CRR286 pKa = 11.84LITKK290 pKa = 10.02GSALPYY296 pKa = 9.82FAAQALQKK304 pKa = 11.23AMWQRR309 pKa = 11.84LQAFPCFKK317 pKa = 10.41LTGCPLDD324 pKa = 4.27ASMLQGLLDD333 pKa = 3.5QEE335 pKa = 4.39EE336 pKa = 4.28RR337 pKa = 11.84LGLDD341 pKa = 3.14FTHH344 pKa = 6.99WVSGDD349 pKa = 3.32YY350 pKa = 10.73SAATDD355 pKa = 3.92GLSQQVNRR363 pKa = 11.84LCLEE367 pKa = 3.9EE368 pKa = 4.7ALRR371 pKa = 11.84GCNASEE377 pKa = 3.81ALKK380 pKa = 10.5IIARR384 pKa = 11.84AVLGNHH390 pKa = 6.64RR391 pKa = 11.84IEE393 pKa = 4.44YY394 pKa = 6.88PTEE397 pKa = 3.9FEE399 pKa = 4.25IDD401 pKa = 3.79GMMQQNGQLMGSPLSFPVLCSINVAAYY428 pKa = 7.28WCALEE433 pKa = 5.16EE434 pKa = 4.0YY435 pKa = 9.38TGRR438 pKa = 11.84KK439 pKa = 8.19FQLEE443 pKa = 4.24DD444 pKa = 3.98LPCLVNGDD452 pKa = 4.86DD453 pKa = 3.24ICFRR457 pKa = 11.84ANPEE461 pKa = 4.4FYY463 pKa = 10.7GIWQRR468 pKa = 11.84WVRR471 pKa = 11.84EE472 pKa = 3.95VGFTLSPGKK481 pKa = 10.4NYY483 pKa = 9.68IHH485 pKa = 6.16EE486 pKa = 4.53HH487 pKa = 5.02FVTVNSEE494 pKa = 4.46GYY496 pKa = 9.0VHH498 pKa = 6.36QKK500 pKa = 9.18HH501 pKa = 5.95GKK503 pKa = 9.21VPFHH507 pKa = 6.71KK508 pKa = 10.96VNFLNTGLLYY518 pKa = 10.57SGKK521 pKa = 9.92SVEE524 pKa = 5.53RR525 pKa = 11.84KK526 pKa = 9.35IDD528 pKa = 3.31WGDD531 pKa = 3.34QPAIKK536 pKa = 10.47VGLRR540 pKa = 11.84PEE542 pKa = 3.97NRR544 pKa = 11.84EE545 pKa = 3.62MPFTQKK551 pKa = 9.85VNRR554 pKa = 11.84VIEE557 pKa = 4.48EE558 pKa = 4.22SCSPTRR564 pKa = 11.84TLLRR568 pKa = 11.84VHH570 pKa = 7.29RR571 pKa = 11.84YY572 pKa = 8.43FRR574 pKa = 11.84DD575 pKa = 3.47EE576 pKa = 4.1IKK578 pKa = 10.77YY579 pKa = 8.06HH580 pKa = 5.13TLNGEE585 pKa = 4.42INMHH589 pKa = 6.9AAPEE593 pKa = 4.15LGGLGIVLPEE603 pKa = 4.37GSDD606 pKa = 3.31TRR608 pKa = 11.84FTAWQCRR615 pKa = 11.84AAGYY619 pKa = 9.02LASRR623 pKa = 11.84WKK625 pKa = 10.44SLEE628 pKa = 3.82FGSVVEE634 pKa = 5.23GIPGPLTQEE643 pKa = 4.08EE644 pKa = 4.34AKK646 pKa = 10.71HH647 pKa = 6.44RR648 pKa = 11.84IVDD651 pKa = 4.05LNRR654 pKa = 11.84PLGVEE659 pKa = 3.95GRR661 pKa = 11.84VTYY664 pKa = 7.93QQKK667 pKa = 10.55KK668 pKa = 8.98SAALTWQAPVKK679 pKa = 9.44PGRR682 pKa = 11.84VVVRR686 pKa = 11.84EE687 pKa = 3.76KK688 pKa = 11.22LEE690 pKa = 3.79PLRR693 pKa = 11.84EE694 pKa = 4.16GEE696 pKa = 4.12EE697 pKa = 4.12RR698 pKa = 11.84IEE700 pKa = 4.42PDD702 pKa = 2.96SSPLRR707 pKa = 11.84NYY709 pKa = 9.9QGPPTSANGEE719 pKa = 4.17WKK721 pKa = 10.35IKK723 pKa = 10.77GLDD726 pKa = 3.61SEE728 pKa = 4.57CRR730 pKa = 11.84QAIRR734 pKa = 11.84EE735 pKa = 4.02YY736 pKa = 10.47RR737 pKa = 11.84GSGVNRR743 pKa = 11.84PLRR746 pKa = 11.84WSRR749 pKa = 11.84EE750 pKa = 3.84VRR752 pKa = 11.84SAPSSILDD760 pKa = 3.3RR761 pKa = 11.84LVRR764 pKa = 11.84SDD766 pKa = 3.47STSVLNTWIGAEE778 pKa = 4.18GQVHH782 pKa = 6.28SFLSKK787 pKa = 10.23PVLVGG792 pKa = 3.35
Molecular weight: 90.01 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
792 |
792 |
792 |
792.0 |
90.01 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.944 ± 0.0 | 2.904 ± 0.0 |
3.662 ± 0.0 | 8.081 ± 0.0 |
3.788 ± 0.0 | 7.449 ± 0.0 |
1.894 ± 0.0 | 4.04 ± 0.0 |
4.293 ± 0.0 | 9.848 ± 0.0 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.146 ± 0.0 | 3.283 ± 0.0 |
5.429 ± 0.0 | 4.545 ± 0.0 |
8.712 ± 0.0 | 6.566 ± 0.0 |
4.545 ± 0.0 | 6.439 ± 0.0 |
2.525 ± 0.0 | 2.904 ± 0.0 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
