
Plasmodium knowlesi (strain H)
Taxonomy: cellular organisms; Eukaryota; Sar; Alveolata; Apicomplexa; Aconoidasida; Haemosporida; Plasmodiidae; Plasmodium; Plasmodium (Plasmodium); Plasmodium knowlesi
Average proteome isoelectric point is 7.09
Get precalculated fractions of proteins
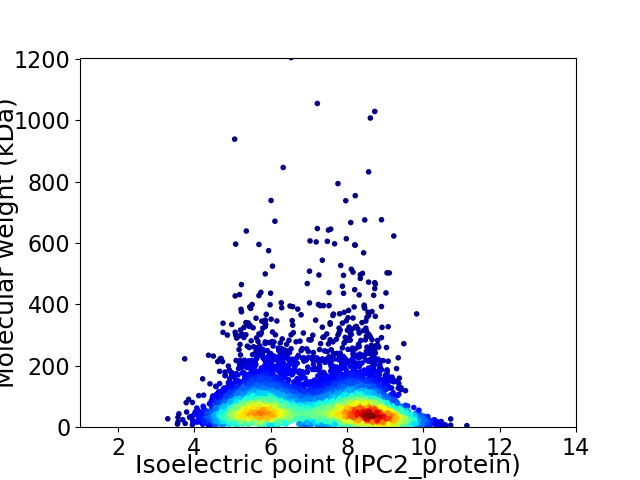
Virtual 2D-PAGE plot for 5339 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
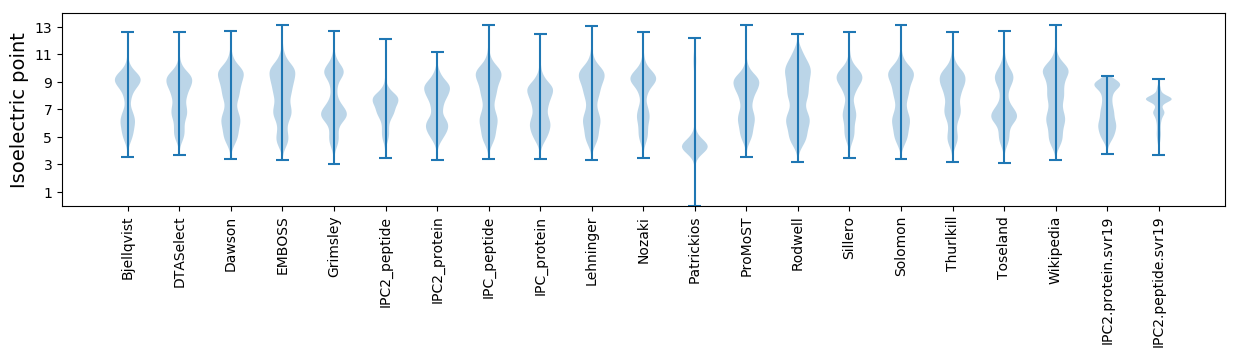
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A679KWN9|A0A679KWN9_PLAKH Uncharacterized protein OS=Plasmodium knowlesi (strain H) OX=5851 GN=PKNH_0838400 PE=4 SV=1
MM1 pKa = 7.82DD2 pKa = 4.35HH3 pKa = 6.41QPPRR7 pKa = 11.84RR8 pKa = 11.84NTNPFIDD15 pKa = 3.9STEE18 pKa = 3.97DD19 pKa = 3.45SINGSDD25 pKa = 3.9TPNVLPVDD33 pKa = 4.89LNSQDD38 pKa = 4.0FGINQWGVGVTEE50 pKa = 4.42GDD52 pKa = 3.76GFSSNVDD59 pKa = 3.23YY60 pKa = 11.93NMGLNSAANGSANGASNGATNGLPNVAPNAAPNAAPNPFANATPNPFANTTPNPFTNTTPNPFTNTTPNPFANTTPNPFANTTPNPFANTTPNPFTNTTPNPFTNTTPNAVPNYY174 pKa = 9.23DD175 pKa = 3.53FNYY178 pKa = 9.1GANSAAQFGQNFNPHH193 pKa = 6.3IGVPSHH199 pKa = 7.06DD200 pKa = 3.78QGNMTSYY207 pKa = 11.01LVDD210 pKa = 2.82STMFNFIQAVVLATTAFLSAVRR232 pKa = 11.84MNRR235 pKa = 11.84HH236 pKa = 4.98MVQPTITKK244 pKa = 10.29NEE246 pKa = 3.47FDD248 pKa = 3.42VAVFHH253 pKa = 6.83NFPTQEE259 pKa = 4.09TTPSGEE265 pKa = 4.12SEE267 pKa = 4.08KK268 pKa = 10.98EE269 pKa = 3.82EE270 pKa = 4.36EE271 pKa = 3.97ISYY274 pKa = 10.91SPLHH278 pKa = 6.51NEE280 pKa = 4.08EE281 pKa = 5.33NNDD284 pKa = 3.88QSDD287 pKa = 3.95TLPLHH292 pKa = 5.76THH294 pKa = 5.72TEE296 pKa = 4.08VHH298 pKa = 6.81PEE300 pKa = 3.96TNTEE304 pKa = 4.16TNTVTNAEE312 pKa = 4.46TNTVTNAEE320 pKa = 4.42TNAEE324 pKa = 4.51TNPEE328 pKa = 4.12IYY330 pKa = 9.97PLLGDD335 pKa = 3.07VDD337 pKa = 4.43YY338 pKa = 11.0EE339 pKa = 4.41HH340 pKa = 7.37EE341 pKa = 4.61PFLPDD346 pKa = 3.59EE347 pKa = 4.56SEE349 pKa = 4.05GDD351 pKa = 3.46IFDD354 pKa = 4.77EE355 pKa = 4.38EE356 pKa = 5.73AEE358 pKa = 4.28DD359 pKa = 4.5SPDD362 pKa = 3.24EE363 pKa = 4.41GEE365 pKa = 4.58YY366 pKa = 10.31EE367 pKa = 4.05VHH369 pKa = 6.31EE370 pKa = 4.6AEE372 pKa = 4.89IQNLMNISYY381 pKa = 9.38DD382 pKa = 3.48IPADD386 pKa = 3.92RR387 pKa = 11.84YY388 pKa = 8.7LTNPFDD394 pKa = 4.03EE395 pKa = 5.45LSNQAGASSGARR407 pKa = 11.84RR408 pKa = 11.84RR409 pKa = 11.84TYY411 pKa = 10.12QPPNYY416 pKa = 10.04GEE418 pKa = 4.32VYY420 pKa = 10.38EE421 pKa = 4.68GAQFVDD427 pKa = 4.59KK428 pKa = 11.12YY429 pKa = 11.21QMSNVGPYY437 pKa = 10.2FINNGNRR444 pKa = 11.84SRR446 pKa = 11.84RR447 pKa = 11.84SGNSEE452 pKa = 4.0SNNYY456 pKa = 9.56GYY458 pKa = 11.46AHH460 pKa = 7.44APYY463 pKa = 10.74ADD465 pKa = 3.74IFRR468 pKa = 11.84HH469 pKa = 5.21PNYY472 pKa = 10.68ANMYY476 pKa = 8.33RR477 pKa = 11.84ATNYY481 pKa = 10.31VMDD484 pKa = 4.57SFLSGTNEE492 pKa = 3.65NTLEE496 pKa = 4.57GIRR499 pKa = 11.84DD500 pKa = 3.84SCSHH504 pKa = 6.43NSADD508 pKa = 4.64DD509 pKa = 3.67SGGANYY515 pKa = 10.48SGGANYY521 pKa = 10.45SGGANYY527 pKa = 10.45SGGANYY533 pKa = 10.67SGDD536 pKa = 3.89DD537 pKa = 3.94NNSGGANYY545 pKa = 10.42SGGANYY551 pKa = 10.62SGDD554 pKa = 3.86ANDD557 pKa = 4.37SGGANYY563 pKa = 10.64SGDD566 pKa = 3.86ANDD569 pKa = 4.37SGGANYY575 pKa = 10.64SGDD578 pKa = 3.86ANDD581 pKa = 4.37SGGANYY587 pKa = 10.64SGDD590 pKa = 3.87ANDD593 pKa = 5.45SGDD596 pKa = 3.7ANNSGGANVSSGSANIGEE614 pKa = 4.16QNIDD618 pKa = 3.05NGYY621 pKa = 10.4DD622 pKa = 3.38DD623 pKa = 5.74SEE625 pKa = 4.44FAHH628 pKa = 6.9TYY630 pKa = 9.28QGNNYY635 pKa = 8.98EE636 pKa = 4.19ITDD639 pKa = 3.87EE640 pKa = 4.95GEE642 pKa = 4.02VADD645 pKa = 4.09PFEE648 pKa = 5.08NVGIDD653 pKa = 3.04NGFDD657 pKa = 3.0FGSYY661 pKa = 9.67EE662 pKa = 4.24NEE664 pKa = 4.02DD665 pKa = 3.88ANGGEE670 pKa = 4.41ATTTTTGSTSQGHH683 pKa = 5.22TVDD686 pKa = 4.91ADD688 pKa = 4.15GITNGDD694 pKa = 3.62VEE696 pKa = 4.88IDD698 pKa = 3.34EE699 pKa = 4.88GDD701 pKa = 3.4NGFGNTEE708 pKa = 3.53KK709 pKa = 10.79HH710 pKa = 5.88EE711 pKa = 4.43PSSEE715 pKa = 3.94KK716 pKa = 10.3TFLTEE721 pKa = 3.88PVKK724 pKa = 10.16NTPTQNDD731 pKa = 4.73TINDD735 pKa = 3.68DD736 pKa = 3.78HH737 pKa = 7.35PSLEE741 pKa = 4.37EE742 pKa = 3.81INVEE746 pKa = 3.96EE747 pKa = 4.35KK748 pKa = 10.78QNSEE752 pKa = 3.99VSEE755 pKa = 4.05QSQPMGHH762 pKa = 7.45PSNDD766 pKa = 3.48DD767 pKa = 3.24DD768 pKa = 6.54DD769 pKa = 5.05EE770 pKa = 5.16FSEE773 pKa = 4.49YY774 pKa = 10.4EE775 pKa = 4.12KK776 pKa = 11.18YY777 pKa = 11.08VYY779 pKa = 8.73TQSGRR784 pKa = 11.84EE785 pKa = 3.91DD786 pKa = 3.09QKK788 pKa = 11.58NYY790 pKa = 9.5NNIMGSYY797 pKa = 8.55LKK799 pKa = 10.26KK800 pKa = 9.85YY801 pKa = 10.47KK802 pKa = 10.13KK803 pKa = 10.29VSEE806 pKa = 4.04NGEE809 pKa = 3.93NEE811 pKa = 3.88KK812 pKa = 11.19AEE814 pKa = 4.21NGVGSDD820 pKa = 3.18AFSPQDD826 pKa = 3.71EE827 pKa = 4.71EE828 pKa = 5.82SPDD831 pKa = 3.41DD832 pKa = 3.75TFPGEE837 pKa = 4.22EE838 pKa = 4.72KK839 pKa = 10.95FNDD842 pKa = 3.77DD843 pKa = 3.64
MM1 pKa = 7.82DD2 pKa = 4.35HH3 pKa = 6.41QPPRR7 pKa = 11.84RR8 pKa = 11.84NTNPFIDD15 pKa = 3.9STEE18 pKa = 3.97DD19 pKa = 3.45SINGSDD25 pKa = 3.9TPNVLPVDD33 pKa = 4.89LNSQDD38 pKa = 4.0FGINQWGVGVTEE50 pKa = 4.42GDD52 pKa = 3.76GFSSNVDD59 pKa = 3.23YY60 pKa = 11.93NMGLNSAANGSANGASNGATNGLPNVAPNAAPNAAPNPFANATPNPFANTTPNPFTNTTPNPFTNTTPNPFANTTPNPFANTTPNPFANTTPNPFTNTTPNPFTNTTPNAVPNYY174 pKa = 9.23DD175 pKa = 3.53FNYY178 pKa = 9.1GANSAAQFGQNFNPHH193 pKa = 6.3IGVPSHH199 pKa = 7.06DD200 pKa = 3.78QGNMTSYY207 pKa = 11.01LVDD210 pKa = 2.82STMFNFIQAVVLATTAFLSAVRR232 pKa = 11.84MNRR235 pKa = 11.84HH236 pKa = 4.98MVQPTITKK244 pKa = 10.29NEE246 pKa = 3.47FDD248 pKa = 3.42VAVFHH253 pKa = 6.83NFPTQEE259 pKa = 4.09TTPSGEE265 pKa = 4.12SEE267 pKa = 4.08KK268 pKa = 10.98EE269 pKa = 3.82EE270 pKa = 4.36EE271 pKa = 3.97ISYY274 pKa = 10.91SPLHH278 pKa = 6.51NEE280 pKa = 4.08EE281 pKa = 5.33NNDD284 pKa = 3.88QSDD287 pKa = 3.95TLPLHH292 pKa = 5.76THH294 pKa = 5.72TEE296 pKa = 4.08VHH298 pKa = 6.81PEE300 pKa = 3.96TNTEE304 pKa = 4.16TNTVTNAEE312 pKa = 4.46TNTVTNAEE320 pKa = 4.42TNAEE324 pKa = 4.51TNPEE328 pKa = 4.12IYY330 pKa = 9.97PLLGDD335 pKa = 3.07VDD337 pKa = 4.43YY338 pKa = 11.0EE339 pKa = 4.41HH340 pKa = 7.37EE341 pKa = 4.61PFLPDD346 pKa = 3.59EE347 pKa = 4.56SEE349 pKa = 4.05GDD351 pKa = 3.46IFDD354 pKa = 4.77EE355 pKa = 4.38EE356 pKa = 5.73AEE358 pKa = 4.28DD359 pKa = 4.5SPDD362 pKa = 3.24EE363 pKa = 4.41GEE365 pKa = 4.58YY366 pKa = 10.31EE367 pKa = 4.05VHH369 pKa = 6.31EE370 pKa = 4.6AEE372 pKa = 4.89IQNLMNISYY381 pKa = 9.38DD382 pKa = 3.48IPADD386 pKa = 3.92RR387 pKa = 11.84YY388 pKa = 8.7LTNPFDD394 pKa = 4.03EE395 pKa = 5.45LSNQAGASSGARR407 pKa = 11.84RR408 pKa = 11.84RR409 pKa = 11.84TYY411 pKa = 10.12QPPNYY416 pKa = 10.04GEE418 pKa = 4.32VYY420 pKa = 10.38EE421 pKa = 4.68GAQFVDD427 pKa = 4.59KK428 pKa = 11.12YY429 pKa = 11.21QMSNVGPYY437 pKa = 10.2FINNGNRR444 pKa = 11.84SRR446 pKa = 11.84RR447 pKa = 11.84SGNSEE452 pKa = 4.0SNNYY456 pKa = 9.56GYY458 pKa = 11.46AHH460 pKa = 7.44APYY463 pKa = 10.74ADD465 pKa = 3.74IFRR468 pKa = 11.84HH469 pKa = 5.21PNYY472 pKa = 10.68ANMYY476 pKa = 8.33RR477 pKa = 11.84ATNYY481 pKa = 10.31VMDD484 pKa = 4.57SFLSGTNEE492 pKa = 3.65NTLEE496 pKa = 4.57GIRR499 pKa = 11.84DD500 pKa = 3.84SCSHH504 pKa = 6.43NSADD508 pKa = 4.64DD509 pKa = 3.67SGGANYY515 pKa = 10.48SGGANYY521 pKa = 10.45SGGANYY527 pKa = 10.45SGGANYY533 pKa = 10.67SGDD536 pKa = 3.89DD537 pKa = 3.94NNSGGANYY545 pKa = 10.42SGGANYY551 pKa = 10.62SGDD554 pKa = 3.86ANDD557 pKa = 4.37SGGANYY563 pKa = 10.64SGDD566 pKa = 3.86ANDD569 pKa = 4.37SGGANYY575 pKa = 10.64SGDD578 pKa = 3.86ANDD581 pKa = 4.37SGGANYY587 pKa = 10.64SGDD590 pKa = 3.87ANDD593 pKa = 5.45SGDD596 pKa = 3.7ANNSGGANVSSGSANIGEE614 pKa = 4.16QNIDD618 pKa = 3.05NGYY621 pKa = 10.4DD622 pKa = 3.38DD623 pKa = 5.74SEE625 pKa = 4.44FAHH628 pKa = 6.9TYY630 pKa = 9.28QGNNYY635 pKa = 8.98EE636 pKa = 4.19ITDD639 pKa = 3.87EE640 pKa = 4.95GEE642 pKa = 4.02VADD645 pKa = 4.09PFEE648 pKa = 5.08NVGIDD653 pKa = 3.04NGFDD657 pKa = 3.0FGSYY661 pKa = 9.67EE662 pKa = 4.24NEE664 pKa = 4.02DD665 pKa = 3.88ANGGEE670 pKa = 4.41ATTTTTGSTSQGHH683 pKa = 5.22TVDD686 pKa = 4.91ADD688 pKa = 4.15GITNGDD694 pKa = 3.62VEE696 pKa = 4.88IDD698 pKa = 3.34EE699 pKa = 4.88GDD701 pKa = 3.4NGFGNTEE708 pKa = 3.53KK709 pKa = 10.79HH710 pKa = 5.88EE711 pKa = 4.43PSSEE715 pKa = 3.94KK716 pKa = 10.3TFLTEE721 pKa = 3.88PVKK724 pKa = 10.16NTPTQNDD731 pKa = 4.73TINDD735 pKa = 3.68DD736 pKa = 3.78HH737 pKa = 7.35PSLEE741 pKa = 4.37EE742 pKa = 3.81INVEE746 pKa = 3.96EE747 pKa = 4.35KK748 pKa = 10.78QNSEE752 pKa = 3.99VSEE755 pKa = 4.05QSQPMGHH762 pKa = 7.45PSNDD766 pKa = 3.48DD767 pKa = 3.24DD768 pKa = 6.54DD769 pKa = 5.05EE770 pKa = 5.16FSEE773 pKa = 4.49YY774 pKa = 10.4EE775 pKa = 4.12KK776 pKa = 11.18YY777 pKa = 11.08VYY779 pKa = 8.73TQSGRR784 pKa = 11.84EE785 pKa = 3.91DD786 pKa = 3.09QKK788 pKa = 11.58NYY790 pKa = 9.5NNIMGSYY797 pKa = 8.55LKK799 pKa = 10.26KK800 pKa = 9.85YY801 pKa = 10.47KK802 pKa = 10.13KK803 pKa = 10.29VSEE806 pKa = 4.04NGEE809 pKa = 3.93NEE811 pKa = 3.88KK812 pKa = 11.19AEE814 pKa = 4.21NGVGSDD820 pKa = 3.18AFSPQDD826 pKa = 3.71EE827 pKa = 4.71EE828 pKa = 5.82SPDD831 pKa = 3.41DD832 pKa = 3.75TFPGEE837 pKa = 4.22EE838 pKa = 4.72KK839 pKa = 10.95FNDD842 pKa = 3.77DD843 pKa = 3.64
Molecular weight: 90.75 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A384LD56|A0A384LD56_PLAKH HSP40 subfamily A putative OS=Plasmodium knowlesi (strain H) OX=5851 GN=PKNH_0424600 PE=4 SV=1
MM1 pKa = 7.28AHH3 pKa = 6.31GASRR7 pKa = 11.84YY8 pKa = 8.75KK9 pKa = 10.48KK10 pKa = 10.3SRR12 pKa = 11.84AKK14 pKa = 9.88MRR16 pKa = 11.84WKK18 pKa = 9.16WKK20 pKa = 9.29KK21 pKa = 9.72KK22 pKa = 6.67RR23 pKa = 11.84TRR25 pKa = 11.84RR26 pKa = 11.84LQKK29 pKa = 9.69KK30 pKa = 7.67RR31 pKa = 11.84RR32 pKa = 11.84KK33 pKa = 8.24MRR35 pKa = 11.84QRR37 pKa = 11.84SRR39 pKa = 3.22
MM1 pKa = 7.28AHH3 pKa = 6.31GASRR7 pKa = 11.84YY8 pKa = 8.75KK9 pKa = 10.48KK10 pKa = 10.3SRR12 pKa = 11.84AKK14 pKa = 9.88MRR16 pKa = 11.84WKK18 pKa = 9.16WKK20 pKa = 9.29KK21 pKa = 9.72KK22 pKa = 6.67RR23 pKa = 11.84TRR25 pKa = 11.84RR26 pKa = 11.84LQKK29 pKa = 9.69KK30 pKa = 7.67RR31 pKa = 11.84RR32 pKa = 11.84KK33 pKa = 8.24MRR35 pKa = 11.84QRR37 pKa = 11.84SRR39 pKa = 3.22
Molecular weight: 5.09 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3890322 |
35 |
10549 |
728.7 |
83.61 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
3.921 ± 0.023 | 1.973 ± 0.019 |
5.778 ± 0.023 | 7.725 ± 0.039 |
4.305 ± 0.029 | 5.644 ± 0.038 |
2.565 ± 0.015 | 6.439 ± 0.034 |
9.698 ± 0.04 | 8.106 ± 0.039 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.324 ± 0.012 | 8.284 ± 0.044 |
3.032 ± 0.021 | 3.246 ± 0.015 |
4.477 ± 0.029 | 7.763 ± 0.029 |
4.739 ± 0.022 | 5.115 ± 0.02 |
0.688 ± 0.01 | 4.177 ± 0.027 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
