
Novosphingobium sp. MBES04
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Sphingomonadales; Sphingomonadaceae; Novosphingobium; unclassified Novosphingobium
Average proteome isoelectric point is 6.2
Get precalculated fractions of proteins
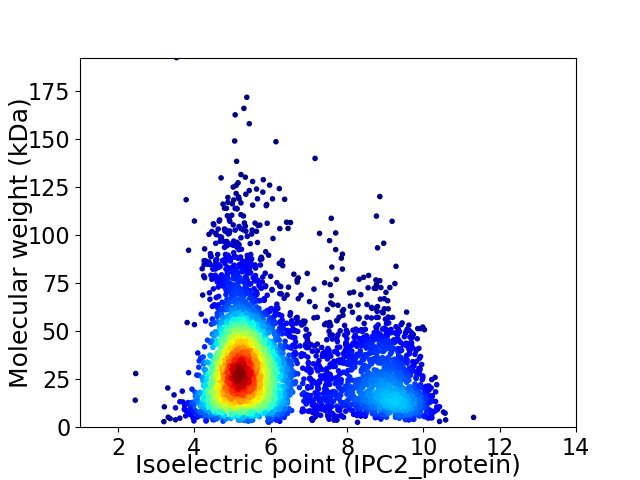
Virtual 2D-PAGE plot for 4731 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
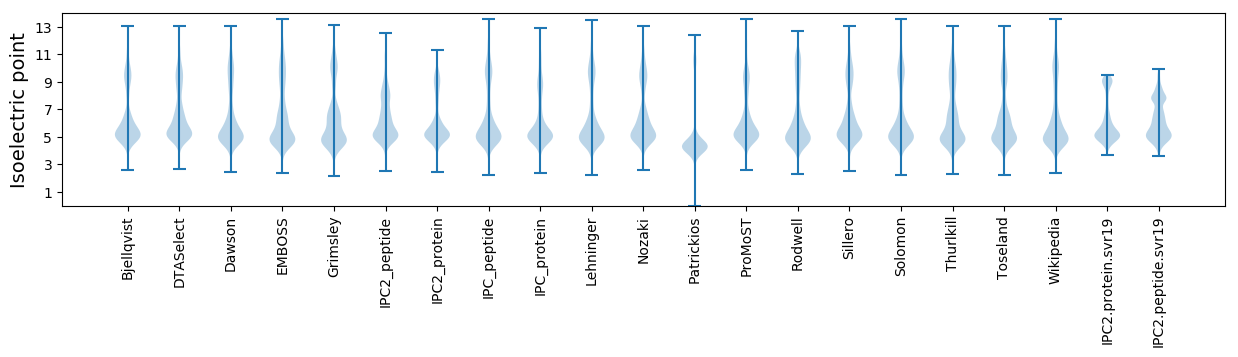
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0S6WWR3|A0A0S6WWR3_9SPHN Regulatory protein RecX OS=Novosphingobium sp. MBES04 OX=1206458 GN=MBENS4_2861 PE=3 SV=1
MM1 pKa = 7.72LLGCSALALAGCGADD16 pKa = 5.41DD17 pKa = 4.13IASPGAGSVVVNNGGGTPTPTPTPTTGIIEE47 pKa = 4.36AAAEE51 pKa = 4.28CPTFNATGGLSNDD64 pKa = 3.69GTIEE68 pKa = 4.21DD69 pKa = 4.57PNGNSWRR76 pKa = 11.84ICTLPALVDD85 pKa = 4.01ASSSLPNEE93 pKa = 3.93AGVLYY98 pKa = 10.16RR99 pKa = 11.84INGRR103 pKa = 11.84VDD105 pKa = 3.16VGCDD109 pKa = 2.96GGFSVPSSGSPYY121 pKa = 7.9TTTTASCLNAGISSLTSDD139 pKa = 3.55TSVEE143 pKa = 3.97LTIDD147 pKa = 3.41PGVIVYY153 pKa = 10.44AEE155 pKa = 4.05NAADD159 pKa = 4.04PAWLAVNRR167 pKa = 11.84GNTIQANGTASSPIVFTSRR186 pKa = 11.84QNVVGSATDD195 pKa = 3.06SSDD198 pKa = 3.66RR199 pKa = 11.84QWGGIVLLGRR209 pKa = 11.84GIITDD214 pKa = 4.03CTAGGSVATDD224 pKa = 3.09DD225 pKa = 4.28CQRR228 pKa = 11.84EE229 pKa = 4.36TEE231 pKa = 4.49GAATPATFGGRR242 pKa = 11.84NNGYY246 pKa = 9.0SAGSMRR252 pKa = 11.84YY253 pKa = 7.87VQIRR257 pKa = 11.84YY258 pKa = 9.22SGYY261 pKa = 10.37NLAPDD266 pKa = 4.93AEE268 pKa = 4.54LQSLTGGGLGTGTTLDD284 pKa = 3.91YY285 pKa = 10.8IQTVNSSDD293 pKa = 3.81DD294 pKa = 3.4GSEE297 pKa = 4.15FFGGAVNMKK306 pKa = 10.27HH307 pKa = 5.88YY308 pKa = 10.43IAVNADD314 pKa = 3.88DD315 pKa = 6.0DD316 pKa = 5.17SLDD319 pKa = 3.46TDD321 pKa = 3.86TGLKK325 pKa = 10.94GNFQYY330 pKa = 11.62LLLLQRR336 pKa = 11.84AGAGDD341 pKa = 3.97AFFEE345 pKa = 4.49IDD347 pKa = 3.75SNNNINSDD355 pKa = 3.47PADD358 pKa = 3.89RR359 pKa = 11.84QRR361 pKa = 11.84STFANFTAIQSGNRR375 pKa = 11.84PDD377 pKa = 4.19NSDD380 pKa = 3.27EE381 pKa = 3.94ASILVRR387 pKa = 11.84GDD389 pKa = 3.03ADD391 pKa = 3.49INFVNGIINTPSNEE405 pKa = 4.26CIRR408 pKa = 11.84VDD410 pKa = 3.07GRR412 pKa = 11.84TATGSATFTANSVVMTCGDD431 pKa = 3.63TGPFLSTGSQYY442 pKa = 11.43SGTNTAANMFNAGTNNNASITSTLTSTFVNGSNEE476 pKa = 4.35SGVVAYY482 pKa = 10.51ANIASFSSFFDD493 pKa = 3.54VVDD496 pKa = 4.92DD497 pKa = 4.21IGAVRR502 pKa = 11.84DD503 pKa = 5.18ANDD506 pKa = 2.47TWYY509 pKa = 10.88RR510 pKa = 11.84GWTCDD515 pKa = 4.03NATADD520 pKa = 3.84FGTGSLCTGLPVAA533 pKa = 5.41
MM1 pKa = 7.72LLGCSALALAGCGADD16 pKa = 5.41DD17 pKa = 4.13IASPGAGSVVVNNGGGTPTPTPTPTTGIIEE47 pKa = 4.36AAAEE51 pKa = 4.28CPTFNATGGLSNDD64 pKa = 3.69GTIEE68 pKa = 4.21DD69 pKa = 4.57PNGNSWRR76 pKa = 11.84ICTLPALVDD85 pKa = 4.01ASSSLPNEE93 pKa = 3.93AGVLYY98 pKa = 10.16RR99 pKa = 11.84INGRR103 pKa = 11.84VDD105 pKa = 3.16VGCDD109 pKa = 2.96GGFSVPSSGSPYY121 pKa = 7.9TTTTASCLNAGISSLTSDD139 pKa = 3.55TSVEE143 pKa = 3.97LTIDD147 pKa = 3.41PGVIVYY153 pKa = 10.44AEE155 pKa = 4.05NAADD159 pKa = 4.04PAWLAVNRR167 pKa = 11.84GNTIQANGTASSPIVFTSRR186 pKa = 11.84QNVVGSATDD195 pKa = 3.06SSDD198 pKa = 3.66RR199 pKa = 11.84QWGGIVLLGRR209 pKa = 11.84GIITDD214 pKa = 4.03CTAGGSVATDD224 pKa = 3.09DD225 pKa = 4.28CQRR228 pKa = 11.84EE229 pKa = 4.36TEE231 pKa = 4.49GAATPATFGGRR242 pKa = 11.84NNGYY246 pKa = 9.0SAGSMRR252 pKa = 11.84YY253 pKa = 7.87VQIRR257 pKa = 11.84YY258 pKa = 9.22SGYY261 pKa = 10.37NLAPDD266 pKa = 4.93AEE268 pKa = 4.54LQSLTGGGLGTGTTLDD284 pKa = 3.91YY285 pKa = 10.8IQTVNSSDD293 pKa = 3.81DD294 pKa = 3.4GSEE297 pKa = 4.15FFGGAVNMKK306 pKa = 10.27HH307 pKa = 5.88YY308 pKa = 10.43IAVNADD314 pKa = 3.88DD315 pKa = 6.0DD316 pKa = 5.17SLDD319 pKa = 3.46TDD321 pKa = 3.86TGLKK325 pKa = 10.94GNFQYY330 pKa = 11.62LLLLQRR336 pKa = 11.84AGAGDD341 pKa = 3.97AFFEE345 pKa = 4.49IDD347 pKa = 3.75SNNNINSDD355 pKa = 3.47PADD358 pKa = 3.89RR359 pKa = 11.84QRR361 pKa = 11.84STFANFTAIQSGNRR375 pKa = 11.84PDD377 pKa = 4.19NSDD380 pKa = 3.27EE381 pKa = 3.94ASILVRR387 pKa = 11.84GDD389 pKa = 3.03ADD391 pKa = 3.49INFVNGIINTPSNEE405 pKa = 4.26CIRR408 pKa = 11.84VDD410 pKa = 3.07GRR412 pKa = 11.84TATGSATFTANSVVMTCGDD431 pKa = 3.63TGPFLSTGSQYY442 pKa = 11.43SGTNTAANMFNAGTNNNASITSTLTSTFVNGSNEE476 pKa = 4.35SGVVAYY482 pKa = 10.51ANIASFSSFFDD493 pKa = 3.54VVDD496 pKa = 4.92DD497 pKa = 4.21IGAVRR502 pKa = 11.84DD503 pKa = 5.18ANDD506 pKa = 2.47TWYY509 pKa = 10.88RR510 pKa = 11.84GWTCDD515 pKa = 4.03NATADD520 pKa = 3.84FGTGSLCTGLPVAA533 pKa = 5.41
Molecular weight: 54.53 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0S6WW08|A0A0S6WW08_9SPHN Pyrimidine monooxygenase RutA OS=Novosphingobium sp. MBES04 OX=1206458 GN=rutA PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.56RR3 pKa = 11.84TFQPSRR9 pKa = 11.84LVRR12 pKa = 11.84ARR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.54GFRR19 pKa = 11.84ARR21 pKa = 11.84TATVGGRR28 pKa = 11.84KK29 pKa = 8.17VLRR32 pKa = 11.84ARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.02KK41 pKa = 10.61LSAA44 pKa = 4.03
MM1 pKa = 7.45KK2 pKa = 9.56RR3 pKa = 11.84TFQPSRR9 pKa = 11.84LVRR12 pKa = 11.84ARR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.54GFRR19 pKa = 11.84ARR21 pKa = 11.84TATVGGRR28 pKa = 11.84KK29 pKa = 8.17VLRR32 pKa = 11.84ARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.02KK41 pKa = 10.61LSAA44 pKa = 4.03
Molecular weight: 5.12 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
1456332 |
26 |
1928 |
307.8 |
33.32 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.005 ± 0.059 | 0.859 ± 0.012 |
5.809 ± 0.027 | 6.056 ± 0.033 |
3.558 ± 0.026 | 8.888 ± 0.035 |
2.035 ± 0.017 | 4.674 ± 0.025 |
2.891 ± 0.026 | 9.965 ± 0.038 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.483 ± 0.018 | 2.508 ± 0.021 |
5.299 ± 0.027 | 3.167 ± 0.02 |
7.175 ± 0.04 | 5.398 ± 0.03 |
5.444 ± 0.026 | 7.032 ± 0.031 |
1.453 ± 0.015 | 2.301 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
